Arabidopsis thaliana gamma-INF induced lysosome thiol reductase gene and application thereof
A reductase, technology of Arabidopsis thaliana, applied in the field of genetic engineering, to achieve strong sulfhydryl reductase activity and wide application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Cloning of AtGILT cDNA
[0027] 1. Extract total RNA from Arabidopsis thaliana.
[0028] Use RNA extraction reagent (TIANGEN) to extract the total RNA of Arabidopsis thaliana according to its operating manual, and identify its purity by formaldehyde denaturing agarose gel electrophoresis, which is greater than 95%, and its concentration measured by ultraviolet spectrophotometer is 720ng / μL.
[0029] 2. Use Transgen cDNA reverse transcription kit to transcribe into first-strand cDNA.
[0030] 1) Primer design: Primers were designed according to the Arabidopsis thaliana database in NCBI and the human GILT sequence alignment: AtGILT-F1: 5'-ATGGCGTCGATGCCGAGCAAACTTC-3', AtGILT-R1 sequence: 5'-TCATAGCAGGCCGGTGATGTCGATG-3'.
[0031] 2) Reverse transcription: Add 3 μL of total RNA extract in sequence to a 1.5 mL Eppendorf tube treated with DEPC, Oligod(T) 18 (0.5μg / μL) 1μL, 2×TS Reaction Mix 10μL, DEPC water 5μL, TransScript RT / RI EnzymeMix 1μL; incubate at 42°C fo...
Embodiment 2
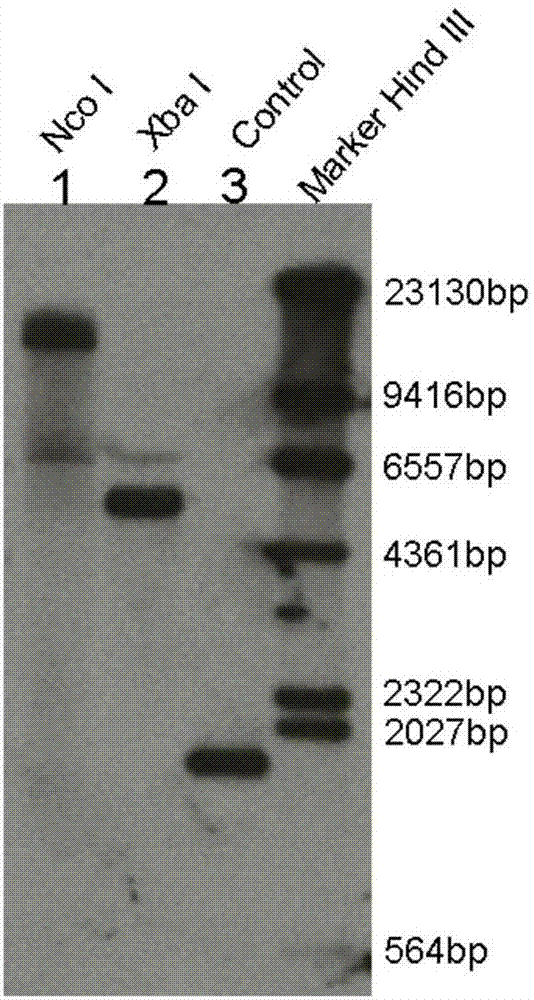
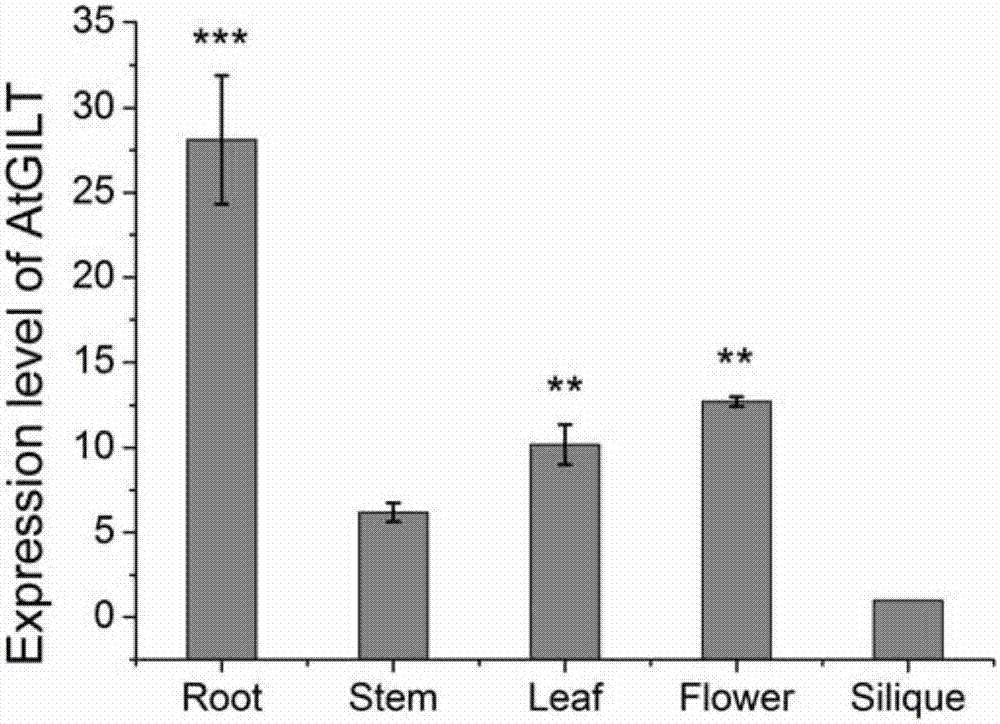
[0033] Gene copy number analysis and expression pattern analysis of embodiment 2AtGILT gene
[0034] 1. First use PCR to prepare probes, using the Arabidopsis genome as a template, the system is 50 μL, 10 μmol / L upstream and downstream primers (upstream primer: 5'-ATCTGATTATTCTGGGGTAT-3'; downstream primer: 5'-TGATGTCGATGTATGACG-3 ') each 1μL, 10×buffer(plus Mg 2+ ) 5 μL, dUTP labeling mixture 5 μL, Taq enzyme 15 μL, ddH2O 36 μL. The reaction program was 94°C for 5 min; 30 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 2 min; finally, an extension at 72°C for 7 min. After the reaction was completed, the product was separated by electrophoresis on a 1% agarose gel, and the DNA band was recovered with a gel extraction kit. Genomic DNA was extracted from Arabidopsis tissue samples and digested with Nco I and Xba I respectively. The system was buffer 60 μL, endonuclease 30 μL, genome 10 μg, ddH 2 O up to 600 μL, digested with an equal volume of phenol: chloroform and extr...
Embodiment 3
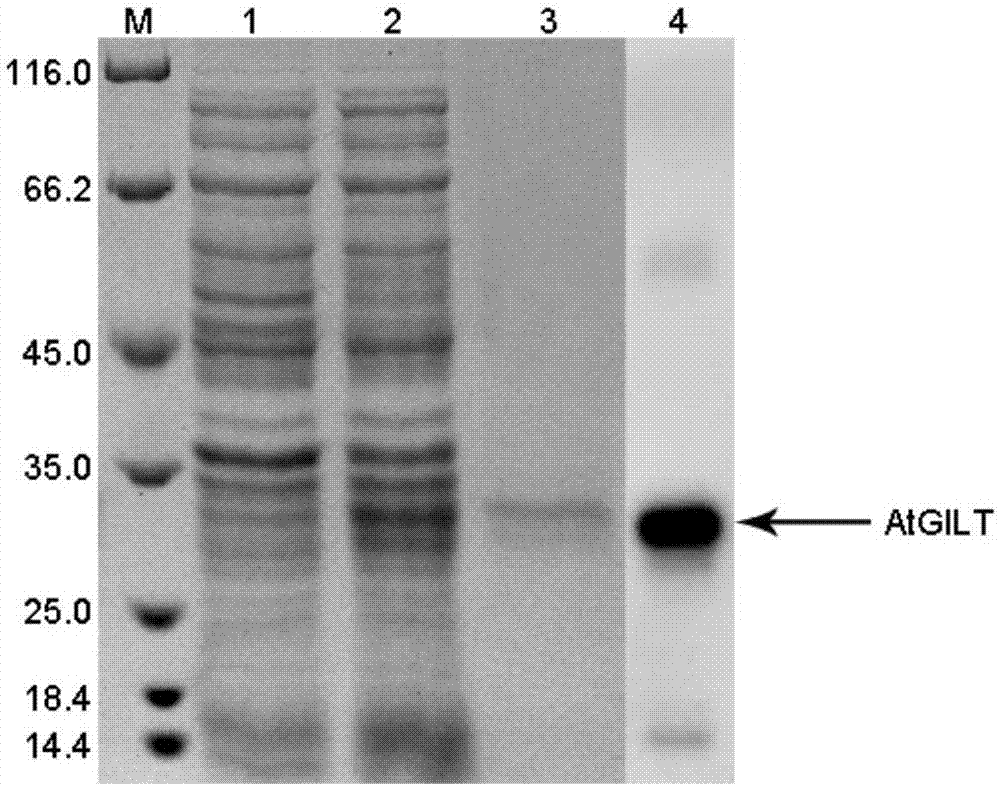
[0036] Example 3 Expression of Arabidopsis thaliana γ-INF-induced lysosomal thiol reductase polypeptide sequence by prokaryotic expression system
[0037] 1. First, use the previously constructed pMD19-T vector connected to the AtGILT gene as a template to perform PCR, the system is 25 μL, 10 μmol / L upstream and downstream primers (upstream primer: 5'-CGGGATCCGATTATTCTGGGGTATCTC-3'; downstream primer: 5'- CCGCTCGAGTCATAGCAGGCCGGTGATG-3') 1μL each, 2.5mmol / L dNTP 2μL, 10×DreamTaqbuffer 2.5μL, cDNA template 1.5μL, DreamTaq enzyme 0.3μL, add ddH 2 O 16.7 μL. The reaction program was: 94°C for 5min; 30 cycles of 94°C for 30s, 58°C for 30s, and 72°C for 1min; finally, an extension at 72°C for 10min. After the reaction was completed, the product was separated by electrophoresis on a 1% agarose gel, and the DNA band was recovered with a gel extraction kit.
[0038] 2. Use BamH I and Xho I to perform double enzyme digestion experiments on the recovered DNA bands. The system is 1 μg ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com