Primer, kit and quantitative detection method for detecting rot valsa mali miyabe et yamada/valsa mali var.dyri
A quantitative detection method and technology for rotting bacteria, applied in the field of fungal molecular detection, can solve the problems of inability to realize biological reaction primers and PCR amplification, and achieve the effects of good primer specificity and avoiding environmental pollution.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Primer Design and Screening
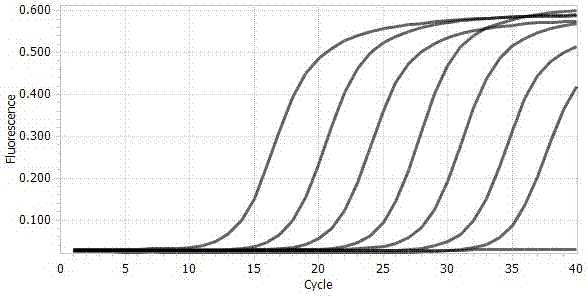
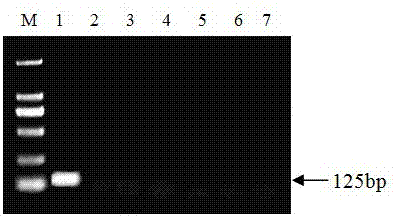
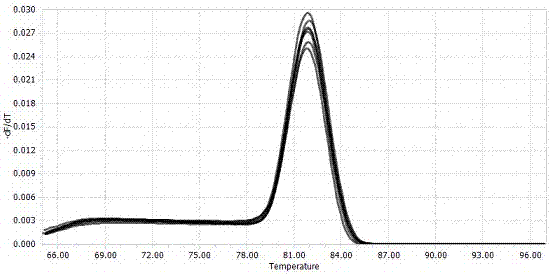
[0037] By comparing the DNA ITS region, β-tubulin gene, and EF-1α elongation factor gene sequences of the apple rot pathogen genome, 13 pairs of primers were designed and synthesized by Shanghai Sangon Biotechnology Co., Ltd. Through specific screening, the specific primer pair VE-F and VE-R were obtained, and the specific amplification effect of the primer pair was as follows: figure 1 shown. It can be seen from the figure that the primers VE-F and VE-R can amplify a clear and single band against the pathogen of apple rot, and the product length is 125 bp. After sequencing and comparison, its sequence is highly consistent with the gene sequence of the pathogen of apple rot. No amplified bands were found for Alternaria, Alternaria, Trichoderma, Fusarium laminarum and Penicillium. By carrying out real-time fluorescent quantitative PCR re-screening on primers VE-F and VE-R, the amplification and melting curves of the apple rot pa...
Embodiment 2
[0040] Example 2 Genomic DNA extraction of apple tree rot fungus 41
[0041] Gently scrape off the mycelium of the apple rot pathogen 41 that just covered the plate on the PDA with a blade, freeze and grind with liquid nitrogen, and extract its DNA by CTAB method: take 0.15 g of the surface-sterilized sample and place it in a 2 mL centrifuge tube. Grind to powder in liquid nitrogen, add 700 μL of extract, and then add 700 μL of phenol:chloroform:isoamyl alcohol mixture, the weight ratio of phenol:chloroform:isoamyl alcohol in the mixture is 25:24:1, shake gently and then centrifuge at 12000 rpm for 15 min, take 400 μL supernatant and add it to a new 1.5 mL centrifuge tube, then add chloroform:isoamyl alcohol mixture with a weight ratio of 24:1 and 40 μLCTAB -NaCl, shake gently and then centrifuge at 12000 rpm for 10 min, take 300 μL of supernatant and add it to another 1.5 mL centrifuge tube, add the same amount of frozen isoamyl alcohol, shake gently and then centrifuge at 12...
Embodiment 3
[0042] Example 3 Acquisition of Positive Recombinant Plasmid Standards
[0043] The DNA of strain 41 was amplified by PCR using primers VE-F and VE-R, and the product was subjected to 1% agarose gel electrophoresis, and the specific band of the target fragment was excised using a DNA gel recovery kit. The gel was recovered, purified and connected to the pUC19 vector, and transformed into Escherichia coli Trans 1-T1 competent cells. The positive clones were picked and cultured, extracted with a plasmid mini-extraction kit, and sequenced for identification. The correctly identified recombinant plasmid was used as a positive recombinant plasmid standard.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com