Grass carp reovirus II type vaccine strain and wild strain diagnostic primer, kit employing diagnostic primer and diagnostic detection method
A technology for differential diagnosis of reovirus, applied in the direction of microorganism-based methods, biochemical equipment and methods, and microbial measurement/inspection, can solve the problems of prevention and control of grass carp haemorrhagic disease, lack of differential diagnosis methods, etc., and achieve Good specificity, closed-tube operation, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Design and synthesis of specific primers
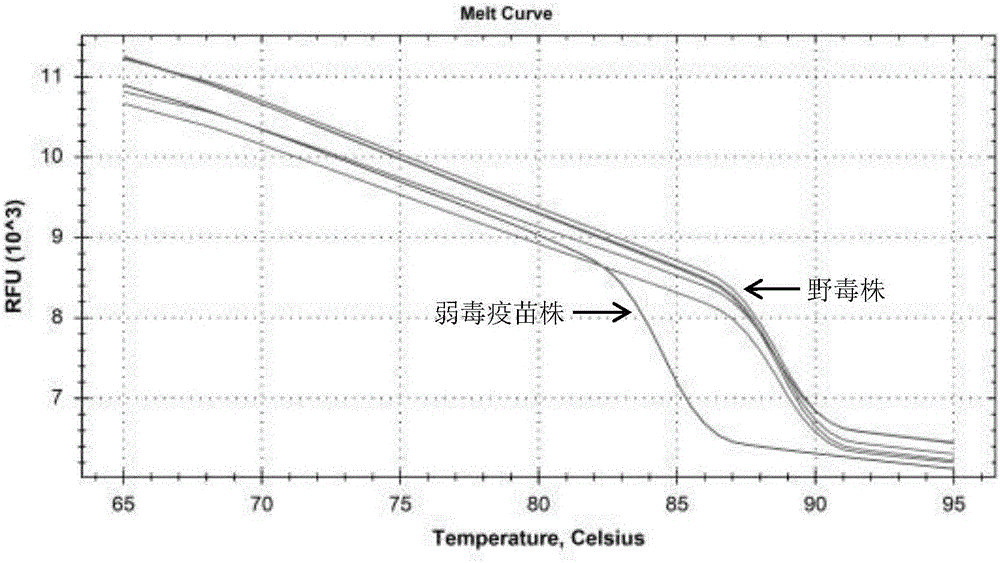
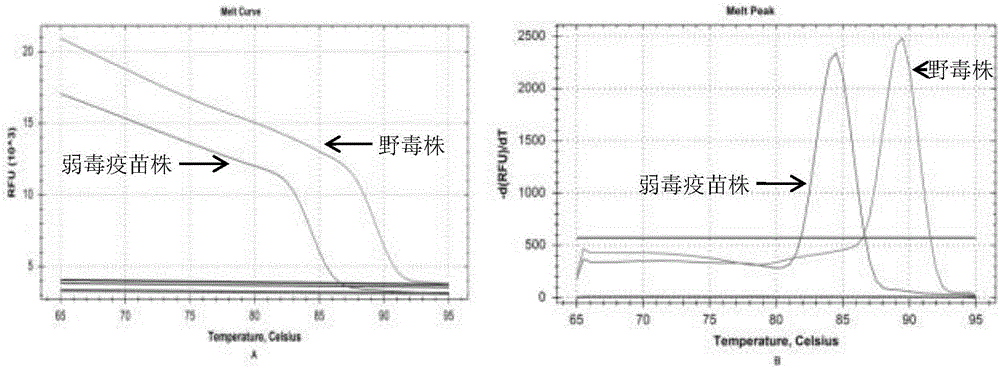
[0036] The S2 gene sequences of 8 GCRV-Ⅱ wild strains and 908 attenuated vaccine strains obtained from NCBI, including HuNan1307, GD108, 106, 918, HeNan988, HeNan794, HZ08, and 109, were compared and analyzed using Oligo 6.0 software, respectively. Primers were designed according to the conserved regions of the 5' and 3' ends of the S2 gene of the grass carp reovirus type Ⅱ vaccine strain and the known type Ⅱ wild strain. base mutation sites. The sequence of the upstream primer HRM-Ⅱ-F is: 5'-CACCGGAGTTAAACTTCATCGC-3', the sequence of the downstream primer HRM-Ⅱ-R is: 5'-CCTGTGGCAGACCGGCAGATAA-3', and the expected length of the PCR amplification product is 233bp.
Embodiment 2
[0037] Example 2: RNA extraction and cDNA preparation
[0038] According to the instructions of the Trizol kit (purchased from Invitrogen, product number: 15596-026) or according to the instructions of the Qiagen RNA extraction kit (purchased from Qiagen, product number: 74104), RNA was extracted from the visceral tissues of fish infected with the corresponding virus. , as a template for RT-PCR reactions. The main steps of Trizol extraction of total RNA are as follows: take the tissue sample and add 500 μl Trizol reagent, mix well and let it stand for 5 minutes, add 400 μl chloroform, shake and mix for 1 minute, centrifuge at 12000 rpm at 4°C for 15 minutes, take the supernatant and add an equal volume of isopropyl Alcohol, mix upside down, place at 4°C for 15min, centrifuge at 12,000rpm at 4°C for 15min, discard the supernatant, wash the pellet with 70% ethanol, centrifuge at 12,000rpm at 4°C for 5min, and resuspend the pellet with 30μl RNAse-free water. The RNA was then rev...
Embodiment 3
[0039] Embodiment 3: GCRVⅡ virus PCR detection
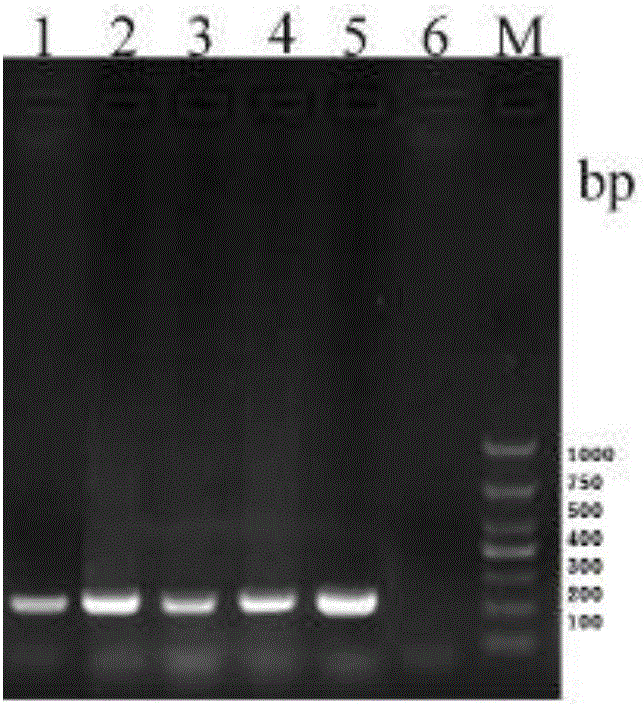
[0040] GSB cells infected by attenuated vaccine strain JX0902 and wild strains HuNan1307, HZ08, GD1607 and GX1407 were detected by PCR method. The PCR system is: cDNA 1.0 μL, upstream primer HRM-Ⅱ-F and downstream primer HRM-Ⅱ-R 0.5 μL, 2.5 mM dNTP 2.0 μL, 10×PCR buffer 1.0 μL, Taq enzyme 0.5 μL, ultrapure water Add to 10 μL. Reaction program: 5min at 95°C; 30s at 95°C, 30s at 55°C, 30s at 72°C; 10min at 72°C; end the reaction at 4°C. Prepare 10g / L agarose electrophoresis gel with agarose and electrophoresis buffer for electrophoresis. After electrophoresis, the gel imaging system was used to observe and collect photos of the electrophoresis gel. The results showed that no matter in the GSB cell samples infected by the attenuated vaccine strain or the wild strain, the band with the same position and a size of about 233bp could be obtained after amplification, which was consistent with the expected result (see figure 1 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com