Primer group for identifying mycoplasma bovis, bovine viral diarrhea virus and infectious bovine rhinotracheitis virus and application thereof
A technique for rhinotracheitis virus and bovine viral diarrhea, which is applied to a primer set for identifying Mycoplasma bovis, bovine viral diarrhea virus and bovine infectious rhinotracheitis virus and its application field, and can solve the problem of indistinguishable, mixed infection, economic loss, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
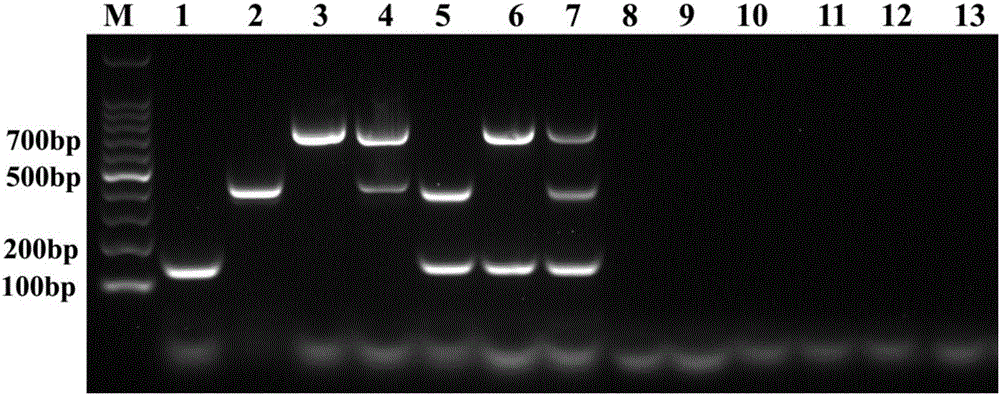
[0077] Embodiment 1, the design of primer
[0078] Sequence analysis was performed on the genomic DNA of Mycoplasma bovis, the DNA corresponding to the total RNA of bovine viral diarrhea virus, and the genomic DNA of bovine infectious rhinotracheitis virus. Based on the sequence analysis, target sites were selected and primers were designed, and thousands of A primer pair library consisting of primer pairs. Preliminary experiments were performed on each primer pair in the primer pair library to test the sensitivity, specificity and universality of the primer pairs, and finally the primer pair A for identifying Mycoplasma bovis and the primer pair A for identifying bovine viral diarrhea virus were obtained. Primer pair B and primer pair C for the identification of bovine infectious rhinotracheitis virus.
[0079] Primer pair A consists of the following primers F1 and R1 (5'→3'):
[0080] F1 (sequence 1 of the sequence listing): GCAACATGAAACCTTATACGA;
[0081] R1 (Sequence 2 ...
Embodiment 2
[0089] Embodiment 2, parameter optimization
[0090] Primer pair A, primer pair B and primer pair C were optimized for triple two-temperature PCR. The parameters to be optimized include parameters of the reaction system and parameters of the reaction program. The parameters of the reaction system include (25 μL): in the initial reaction system, the content of AMV reverse transcriptase, MgCl 2 The concentration of Taq DNA Polymerase, the concentration of dNTP, and the concentration of each primer. Parameters of the reaction program include: annealing extension temperature.
[0091] The recommended initial reaction system is (25 μL): AMV reverse transcriptase 1~5U, MgCl 2 1~10mmol / L, Taq DNA Polymerase 1~5U, dNTP 0.1~0.8mmol / L, each primer 1~10pmol / μL. The optimal initial reaction system is (25μL): AMV reverse transcriptase 5U, MgCl 2 1.5mmol / L, Taq DNA Polymerase 2.5U, dNTP 0.2mmol / L, primer F1 1pmol / μL, primer R1 1pmol / μL, primer F2 1pmol / μL, primer R2 1pmol / μL, primer ...
Embodiment 3
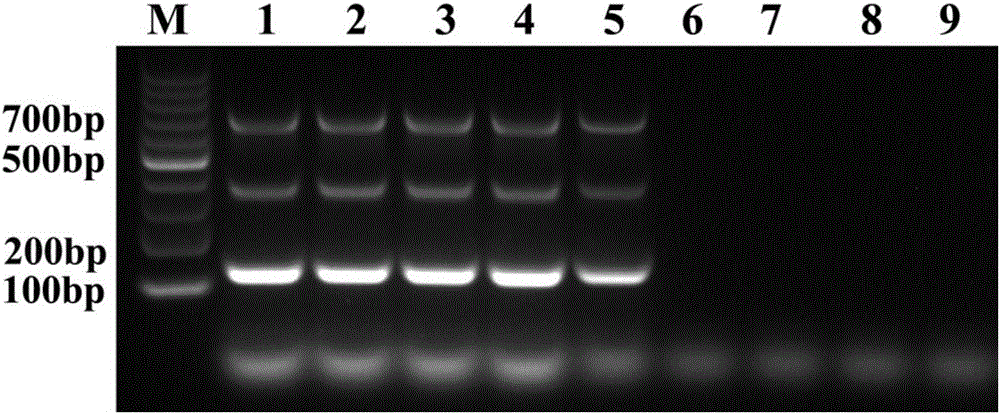
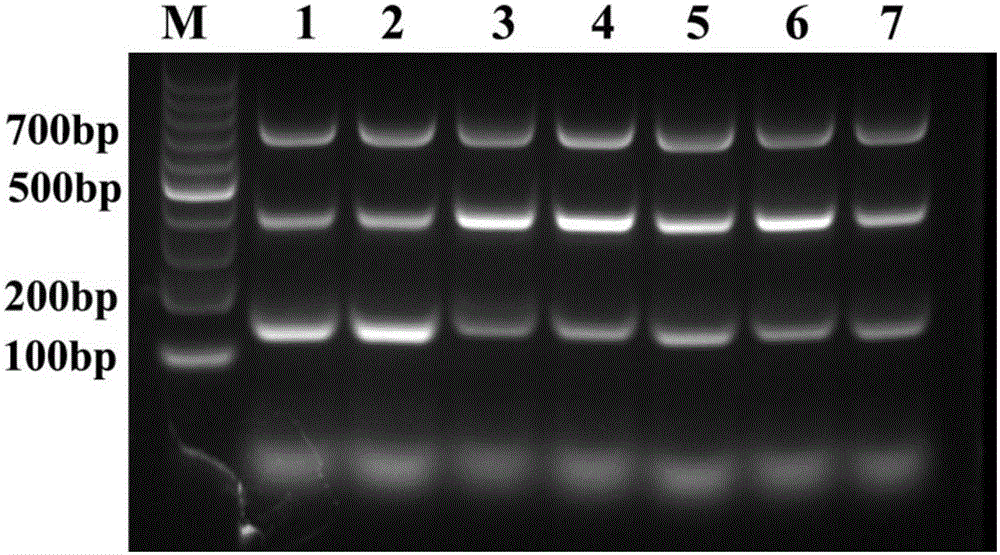
[0093] Embodiment 3, establishment of method
[0094] 1. Use the RNA / DNA co-extraction kit to extract the nucleic acid of the sample to be tested.
[0095] 2. Take the nucleic acid obtained in step 1 as a template and perform triple two-temperature PCR.
[0096] The initial reaction system is (25μL) AMV reverse transcriptase 5U, MgCl 2 1.5mmol / L, Taq DNA Polymerase2.5U, dNTP 0.2mmol / L, Primer F1 1pmol / μL, Primer R1 1pmol / μL, Primer F2 1pmol / μL, Primer R2 1pmol / μL, Primer F3 1pmol / μL, Primer R3 1pmol / μL μL, 2.5 μL of 10×buffer, 1 μL of template, and water as the balance.
[0097] The reaction program is: 42°C for 30 minutes; 94°C for 5 minutes; 94°C for 30s, 67°C for 30s, 35 cycles; 72°C for 10 minutes.
[0098] 3. Take the product of step 2 and perform 1% agarose gel electrophoresis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com