Method for constructing negative bacteria capable of expressing polysaccharides of positive bacteria and mutants thereof
A technology for mutants and Gram-positive bacteria, applied in the field of bioengineering, can solve problems such as polysaccharides of Gram-positive bacteria that have not yet been discovered, and achieve the effects of promoting research and development, easy cultivation, and easy genetic engineering transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
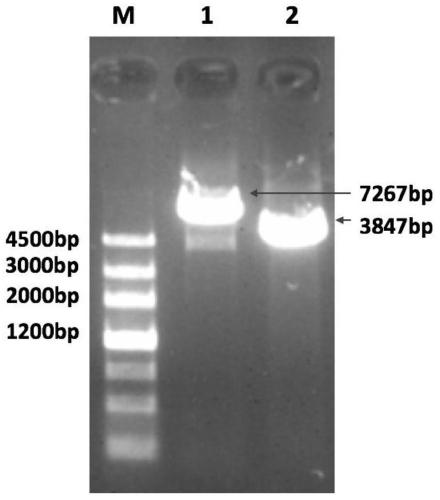
[0043] Two pairs of PCR primers with overlapping fragments were designed according to the reported sequence of the whole gene cluster strain of capsular polysaccharide of Streptococcus pneumoniae serotype 6A serotype (refer to GenBank sequence number GCA_001088685.1) to classify the full-length gene cluster with a full length of 7267 bp. The two segments were amplified, and the sizes of the amplified fragments were 3420bp and 3847bp respectively. The above primers were synthesized by Beijing Huada Gene Company. The primer sequences are as follows:
[0044] 6A-1F: 5'-TCACACAGGAAACAGACCATGAATGGAAAAATAGTAAAG-3'
[0045] 6A-1R: 5'-TGTTTGAGACCTAATACTATC-3'
[0046] 6A-2F: 5'-TATAATGGTGAGCGATATTTG-3'
[0047] 6A-2R: 5'-GTTATTGTCTCATGAGCGTTATTCAAATATTTTCTTTCT-3'
[0048] At the same time, it is necessary to use the vector pYA3337 as a template to design a pair of PCR primers with overlapping fragments to amplify the vector fragment. The primers were synthesized by Beijing Huada Gen...
Embodiment 2
[0062] LPS pattern identification: for the mutant strain of embodiment 1, cultivate overnight 5ml PCR and identify as the positive Salmonella typhimurium strain that contains expression plasmid, centrifuge and collect thalline, get 200 μ l buffer A (component is: 0.5M Tris-Cl pH 6.8, 10% glycerol, 10% SDS and 5% Beta-mercaptoethanol) resuspended bacteria, the sample was thoroughly mixed and boiled in boiling water for 10 minutes, after the sample was cooled, centrifuged for 15 minutes to remove undissolved impurities, centrifuged After completion, add the supernatant to buffer B (components: 0.5M Tris-Cl pH 6.8, 10% glycerol, 0.05% Bromophenol blue) at a ratio of 1:10 (10 μl to 90 μl), and add 1 μl concentration 20 mg / ml proteinase K. After mixing well, place the sample at 37°C. One hour later, take 15 μl of the sample for electrophoresis on a 15% SDS-PAGE gel. After the gel is run, the gel is stained with silver ammonia. If the capsular polysaccharide of Streptococcus pneumo...
Embodiment 3
[0069] Western blot identification: the processed lipopolysaccharide samples were separated by SDS-PAGE gel, and after the electrophoresis was completed, the LPS band was transferred to a nitrocellulose membrane by a transfer machine. After the transfer was completed, it was blocked with Tris buffer containing 5% skimmed milk for 2 hours, then rabbit antiserum (diluted 1:100) specific for Streptococcus pneumoniae serotype 6A capsular polysaccharide was added and incubated at room temperature for 2 hours. After the incubation, the goat anti-rabbit IgG secondary antibody labeled with alkaline phosphatase was incubated at a 1:10000 dilution, and washed three times with TBS buffer two hours later. The immunoblotting bands were developed by adding BCIP color developing solution. The reaction is terminated by washing the membrane in ultrapure water to terminate the reaction. Salmonella typhimurium without plasmid was used as negative control. If there is a band, it means that the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com