Method for detecting kanamycin
A kanamycin and detection method technology, applied in the field of sensors, can solve problems such as complex instrument operation, and achieve the effects of improving sensitivity, mild reaction conditions and accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
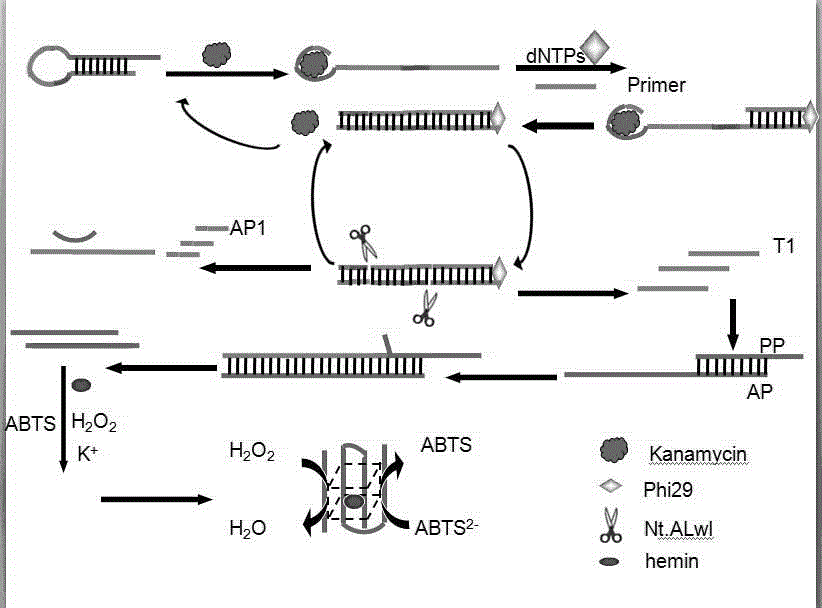
[0038] The preparation method of described biosensor comprises the following steps:
[0039] (1) Specific binding of target and hairpin;
[0040] (2) Amplify the signal of the detected specific sequence;
[0041] (3) Perform colorimetric detection on the sequence.
[0042] In the preparation method, it is preferable to react the target substance with the hairpin, and the operation steps are as follows: mix 2 μL of the target substance kanamycin with 2 μL of the hairpin, and incubate at 37° C. for 0.5 h.
[0043] In the preparation method, the process of specifically amplifying the detected signal is preferably as follows:
[0044] (2) Take out the mixed solution incubated in (1), add primers, dNTPs, phi29DNA polymerase and Nt.Alwi endonuclease to the mixed solution, shake for 30s, and incubate in a 37°C incubator for 1h;
[0045] In the preparation method, the addition of the enzyme is carried out at low temperature to ensure the activity of the enzyme.
[0046] (3) Mix th...
Embodiment 1
[0051] The main steps of probe hybridization are as follows:
[0052] a. Add 2 μL of PHI29 buffer to the centrifuge tube (buffer is the supporting buffer for purchasing PHI29 DNA polymerase);
[0053] b. Add 8 μL of sterilized water into the centrifuge tube and shake evenly;
[0054] At this time, we will melt the chain again:
[0055] a. Add in order according to the number of sterilized water required to dissolve one O.D corresponding to the HAP chain;
[0056] b. Put the probe chain added with sterilized water into the centrifuge, set at 3000r / min, and centrifuge for 30 seconds;
[0057] c. Take out the centrifuged HAP chain, add 2 μL of kanamycin, tetracycline, and streptomycin to each centrifuge tube, and shake evenly.
[0058] d. Add 2 μL primers, 2 μL dNTPs, 2 μL phi29 DNA polymerase and 2 μL Nt.Alwi endonuclease to each centrifuge tube in turn, and incubate in a 37-degree incubator for one hour;
[0059] e. Mix 2 μL AP and 2 μL PP and add to each centrifuge tube, a...
Embodiment 2
[0062] The main steps of restriction enzyme optimization are as follows:
[0063] a. Add 2 μL of DNTP and 2 μL of Phhi29 DNA polymerase to the centrifuge tube where the target substance was added, and then add 1 μL of Nt.Alwi endonuclease, 1.5 μL of Nt.Alwi endonuclease, 2 μL of Nt.Alwi endonuclease and 2.5 μL of Nt to each centrifuge tube .Alwi endonuclease, 3μL Nt.Alwi endonuclease;
[0064] b. After shaking evenly, incubate at 37°C for 1 hour;
[0065] The following describes the reaction that occurs in a homogeneous solution, and the main steps in a homogeneous reaction:
[0066] c. Add sterilized water, 10× buffer, HAP (2 μL), kanamycin (2 μL), phi29 DNA polymerase (2 μL), dNTPs (2 μL) into the centrifuge tube, and shake for 30 seconds.
[0067] d. Add different concentrations of Nt.Alwi endonuclease to the mixed solution in the centrifuge tube in turn;
[0068] e. Incubate the mixed solution in (d) in an incubator at 37°C for 1 hour.
[0069] f. Add the hybrid soluti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com