Oxidation hydrolase gene BtLPMO10A and oxidation hydrolase and application
A technology of hydrolase and gene, which is applied in the field of preparation of oxidative hydrolase, can solve the problems that the crystal structure is difficult to be destroyed, and the catalytic efficiency of glycoside hydrolase is low, so as to achieve the effect of large production potential and economic value, and improve the degradation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 The cultivation of Bacillus thuringiensis subspecies Kustak and the extraction of its genomic DNA
[0025] A single clone of Bacillus thuringiensis subsp. Kustak strain (purchased from the China Agricultural Microorganism Culture Collection and Management Center, No. ACCC10066) was inoculated into 10ml liquid LB medium, and then placed in a temperature of 30°C and a rotation speed of 150rpm Cultivate on a shaker for 16 hours, take 3ml of the bacterial liquid, and centrifuge to collect the bacterial cells for the extraction of genomic DNA. The extraction and purification methods of genomic DNA were completed according to the instructions of the kit.
Embodiment 2B
[0026] Recombinant expression of embodiment 2BtLPMO10A gene in Escherichia coli
[0027] Using the genomic DNA of Bacillus thuringiensis subspecies Kustak ACCC10066 as a template, PCR amplification was performed with the following primer pairs. Primers were designed as follows:
[0028] Forward primer P-F: 5'-cgctgcccagccggcgatggcccatggatatgtagaatcaccagcg-3';
[0029] Reverse primer P-R: 5'-gctttgttagcagccggatctcaaaatagtgtaggagtttgcatacg-3'; polymerase PrimeSTAR was purchased from Treasure Biotech, and the PCR reaction system was operated according to the product instructions provided by the company. PCR reaction conditions: 94°C pre-denaturation for 5 minutes, then denaturation at 94°C for 30 sec-50°C annealing for 30 sec-72°C extension for 1 min, 30 cycles, and finally 72°C extension for 10 min. After the PCR reaction, the PCR product was used as a primer, and the pET22b plasmid was used as a carrier to carry out secondary PCR amplification according to the above PCR ampli...
Embodiment 3
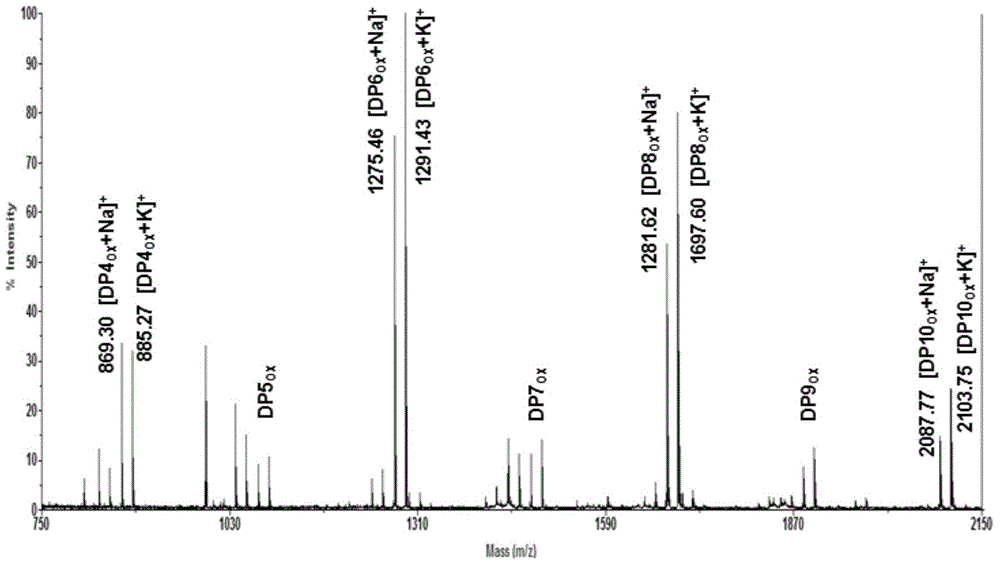
[0057] Example 3 Analysis of biochemical properties of oxidohydrolase BtLPMO10B
[0058] (1) Analysis of polysaccharide binding ability
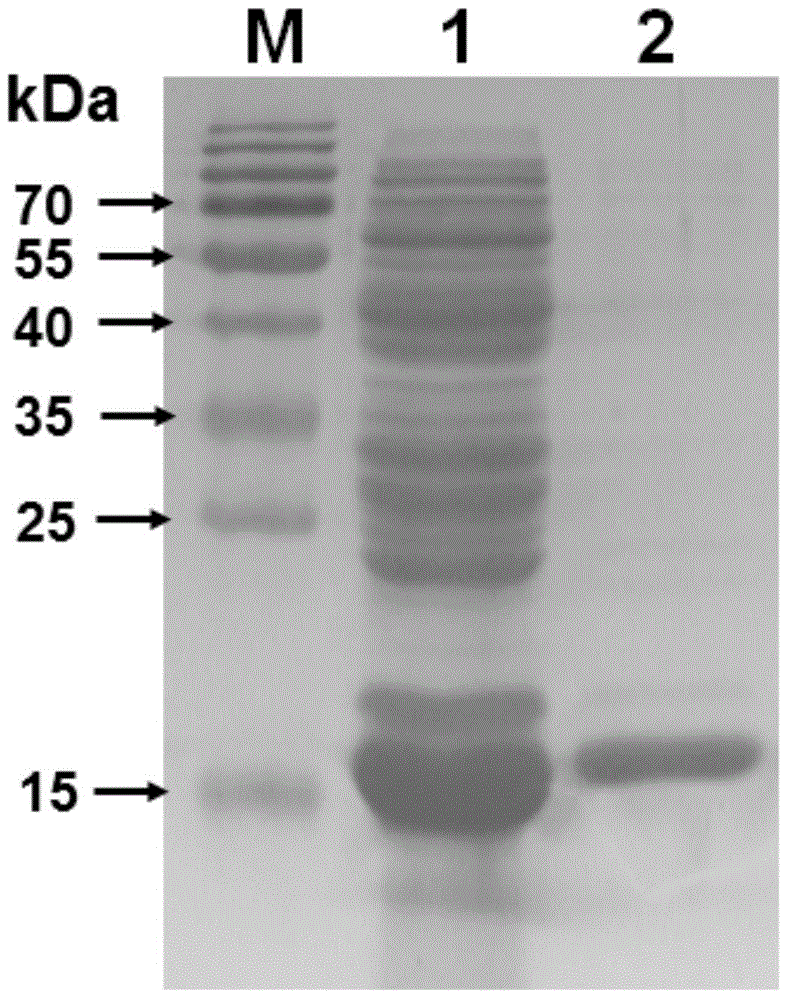
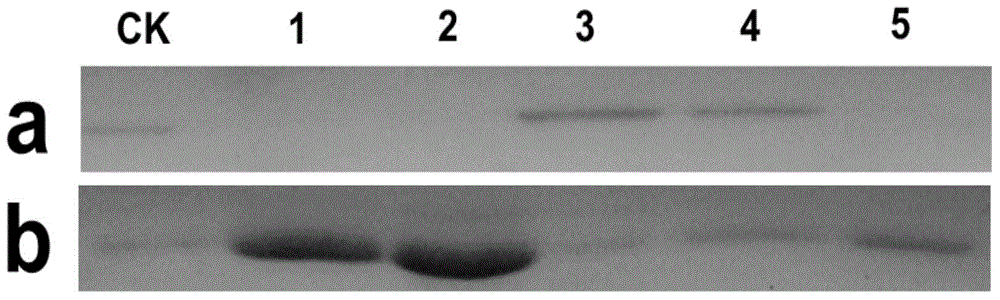
[0059] The binding experiment between oxidohydrolase BtLPMO10A and various polysaccharides was carried out according to the following conditions: 5 mg of various polysaccharides were mixed with 0.5 mg of purified oxidohydrolase BtLPMO10A in a 1.5 ml Eppendorf tube, and the final volume of the reaction was passed through 20 mM, pH 8.0 Make up to 1 ml of Tris-HCl buffer. The combination of enzyme and substrate was carried out at room temperature for 24 hours. In order to make the combination of enzyme and polysaccharide better, a DH-II rotary mixer (Ningbo Xinzhi Company) was used. After the end, the supernatant and the precipitate were collected by centrifugation, and then detected by SDS-PAGE electrophoresis. The result is as figure 2 As shown, the oxidohydrolase BtLPMO10A has a strong binding ability to crystalline chitin (α-chitin and ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com