Application of serum microRNA in diagnosis of liver cancer and diagnostic kit
A diagnostic kit, hsa-mir-192 technology, applied in the determination/testing of microorganisms, biochemical equipment and methods, etc., can solve the problem of inability to comprehensively and objectively reflect circulating microRNA, difficult to explain marker diagnostic markers, and lack of multi-center Validation and other issues to achieve the effect of saving time and labor costs, mature experimental methods, and easy access
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Collection and preparation of serum samples
[0056] The inventor collected healthy people (HC), hepatitis B carriers (IHC), chronic hepatitis B patients (CHB), liver cirrhosis patients (LC) and liver cancer patients (HCC) from August 2009 to August 2014. These populations met the inclusion criteria (Table 1), and according to the principle of gender and age matching, liver cancer and its control group specimens were set.
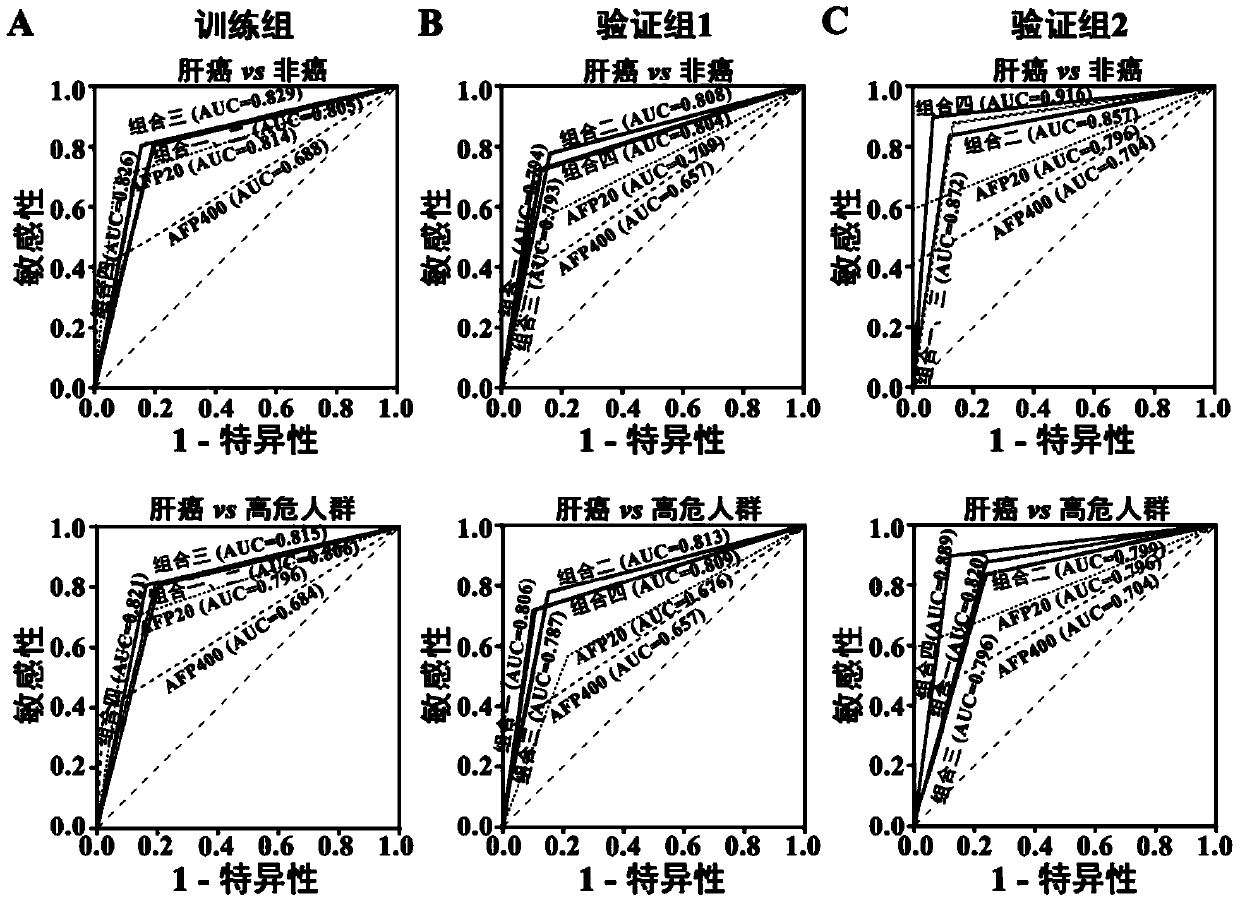
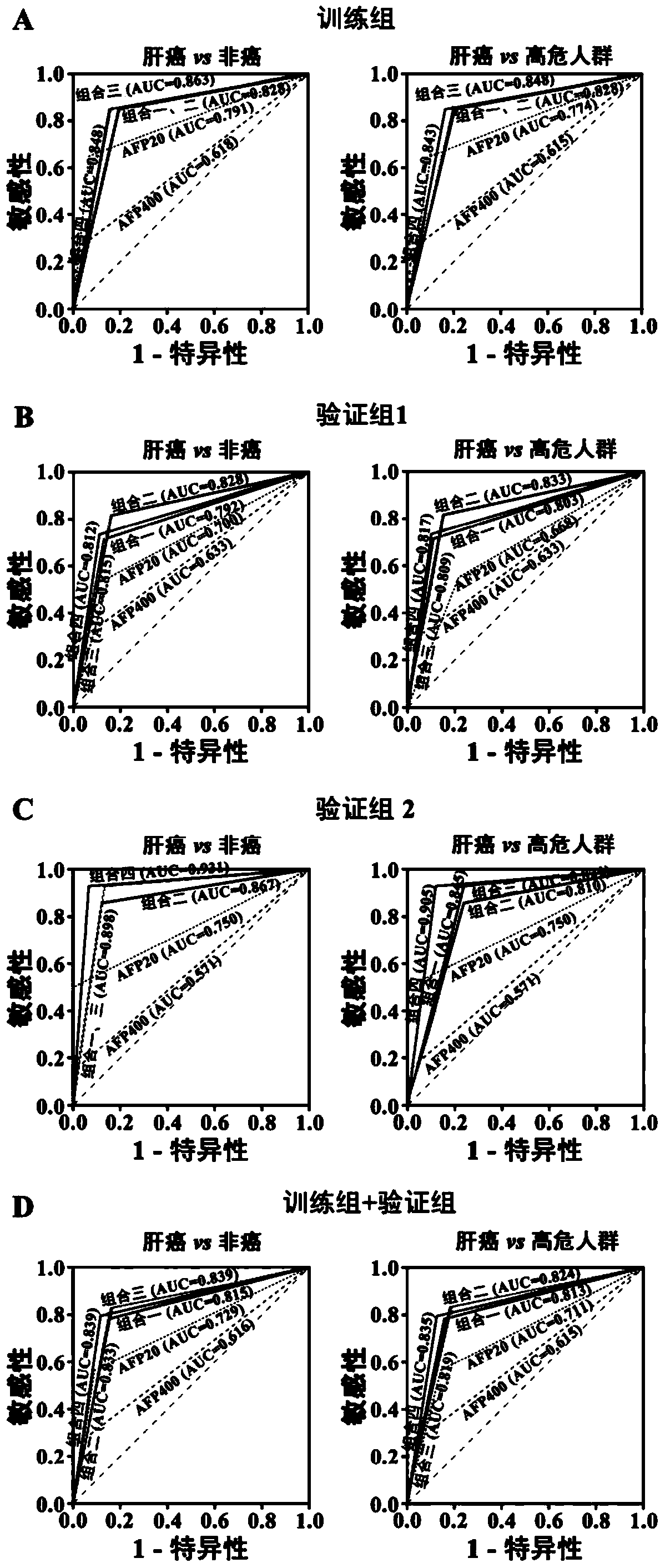
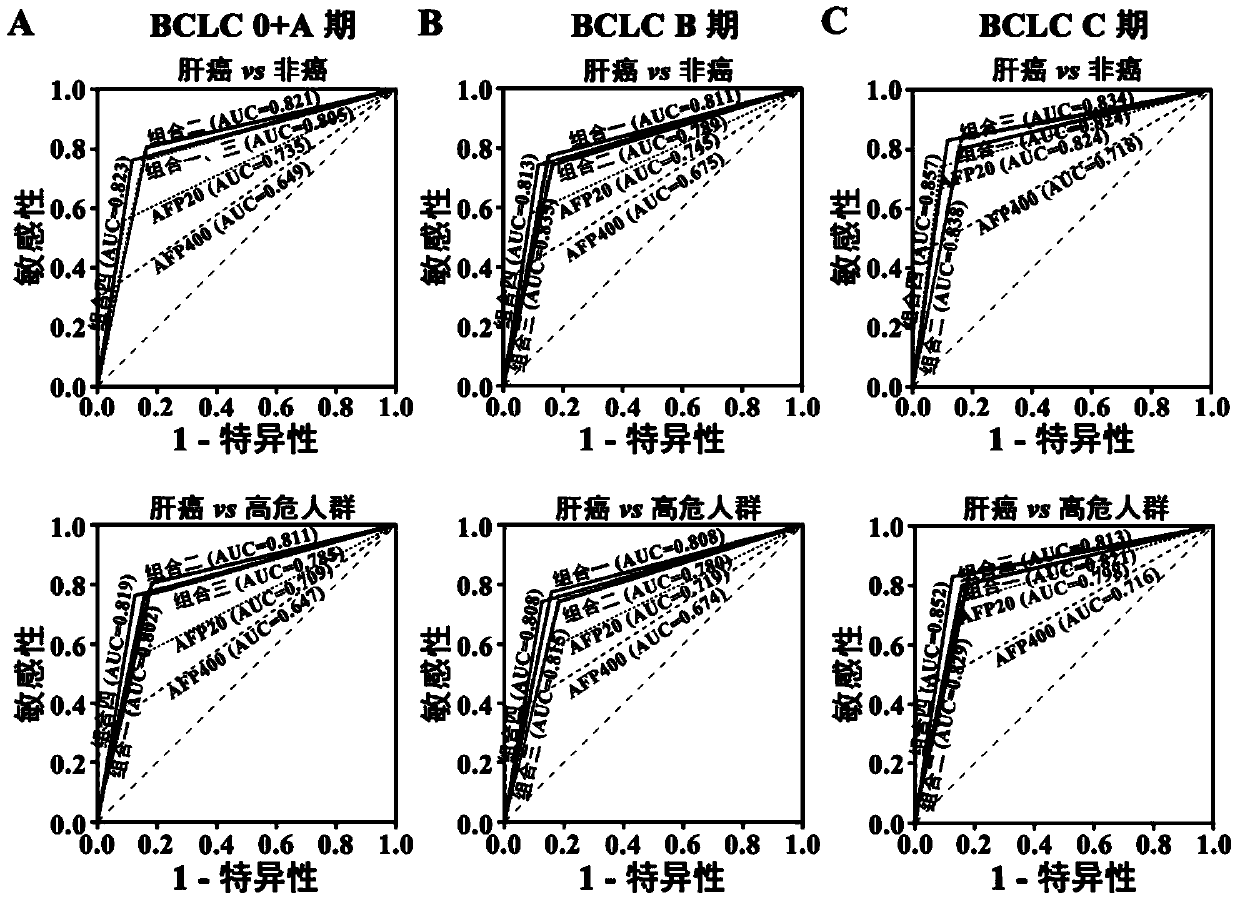
[0057] Training group: 257 serum samples collected from healthy people (51 cases), chronic hepatitis B patients (51 cases), liver cirrhosis patients (47 cases) and liver cancer patients (108 cases) from August 2009 to March 2012 .
[0058] Validation group 1: 352 serum samples from healthy people (60 cases), chronic hepatitis B patients (68 cases), liver cirrhosis patients (71 cases) and liver cancer patients (153 cases) collected from April 2012 to April 2013 specimen.
[0059] Validation group 2: 139 serum samples from healthy people ...
Embodiment 2
[0068] Example 2: qPCR Array and its data analysis
[0069] Serum samples from 6 patients with liver cancer before operation and 8 patients with chronic hepatitis B at least one year before the last examination were selected for qPCR Array screening. These patients were all male, and there was no significant difference in mean age and distribution (Table 3).
[0070] Table 3. Clinicopathological characteristics of specimens used for qPCR Array analysis
[0071]
[0072] The present invention adopts Applied Biosystems company's The Array Human MicroRNA method screens microRNAs that differ between liver cancer and chronic hepatitis B, and detects the levels of 754 known human microRNAs. See the Applied Biosystems website for specific steps. After the original data obtained were calibrated, the Significant Analysis of Microarray (SAM) analysis method was used to select differential microRNAs, and finally 19 candidate microRNAs for subsequent verification were screened (Tab...
Embodiment 3
[0076] Example 3: Real-time fluorescence quantitative PCR detection training group specimen microRNA level
[0077] 1.1 Extraction of serum RNA
[0078] The present invention uses Trizol reagent to extract, and obtains serum RNA through the method of phenol / chloroform extraction and purification, isopropanol precipitation, and glycogen-assisted precipitation. The specific steps are as follows:
[0079] 1. Take 200 μl of serum, preferably add an equal volume of double-stranded RNA mixed with cel-miR-67 (NC67, based on the mature body sequence of nematode miR-67 that has no homology with the human genome sequence, and the final concentration is 0.2 nM, the sequence is shown in Table 4) of Trizol lysate, fully vortexed and mixed, and ice-bathed for 15 minutes.
[0080] 2. Add 100 μl of pre-cooled chloroform, shake and mix well, and centrifuge at 12,000 g for 15 min at 4°C.
[0081] 3. Transfer the supernatant, add an equal volume of pre-cooled phenol / chloroform (1:1), shake and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com