Method for knocking out fungus genes
A gene knockout and fungal technology, applied in the field of genetic engineering, can solve the problems of few types and limitations of site-specific DNA recombinases, and achieve the effect of avoiding the effect of SCRaMbLE and improving the stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1 Contains the construction of the recombinant bacterial strain of Vika-vox recombinant system
[0059] Experimental Materials:
[0060] Saccharomyces cerevisiae codon optimization was performed according to the reported amino acid sequence of Vika recombinase. The DNA molecule encoding Vika recombinase after optimization has the nucleotide sequence shown in SEQ ID NO: 3, and the gene was synthesized in Jinweizhi Company to obtain plasmid pUC57-Vika .
[0061] According to the reported CreEBD fusion gene with SCW11 promoter, which has the nucleotide sequence shown in SEQ ID NO: 24, the gene was synthesized in Jinweizhi Company to obtain the plasmid pUC57-pSCW11-CreEBD.
[0062]The strain Saccharomyces cerevisiae synIII, which artificially synthesized the third chromosome of Saccharomyces cerevisiae, was donated by the Jef Boeke laboratory of Johns Hopkins University in the United States, and its chromosome sequence is numbered in GenBank: KC880027.1 (http: / / ...
Embodiment 2
[0079] Example 2: Knockout of Saccharomyces cerevisiae Chromosomal Fragments Using Vika-vox Recombinase System
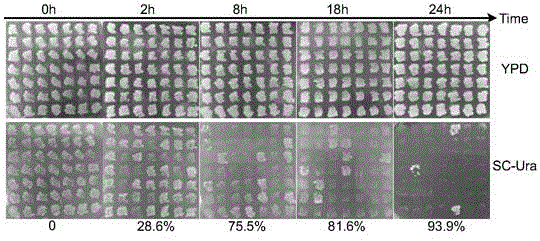
[0080] The recombinant strain yLQH207 was inoculated into the medium for cultivation. After the glucose in the medium was exhausted, galactose mother solution (20%) was added to make the final concentration of galactose 20g / L to induce the expression of the GAL1 promoter. The bacterial solution after 0h, 2h, 8h, 18h and 24h of induction with galactose was selected and spread on the YPD plate, and cultured at 30°C for 2 days. 49 single colonies were randomly picked and streaked to the YPD plate, cultured at 30°C for 24 hours, and then copied to the SC-Ura plate. Because the length of the fragment between the two vox on the yLQH207 genome is 6.2kb, when the Vika-vox recombinase system is activated for gene knockout, its phenotype is changed by Ura + Trp - becomes Ura - Trp - , leaving a scar sequence of 34bp vox site on the genome. Since there is a Ura3 nutrient ...
Embodiment 3
[0084] Embodiment 3: Utilize Vika-vox recombination system to knock out the application of genome Cre gene
[0085] (1) Construction of Cre recombinant strains
[0086] Using plasmid pRS414 as template and trp1L-vox-F / R as primers, the genome integration fragment trp1L-vox (385bp) was amplified. Using the plasmid pUC57-pSCW11-CreEBD as a template and pSCW11-Cre-F / R as a primer, the genome integration fragment pSCW11-CreEBD-2 (3443bp) was amplified. Using plasmid pRS416 as template and Ura3-Cre-F / Ura-vox-R as primers, PCR was carried out to obtain the genome integration fragment Ura3. Using plasmid pRS414 as template and vox-Trp1R-F / R as primers, PCR was carried out to obtain the genome integration fragment vox-Trp1R (390bp). The above four genome integration fragments with homology arms were obtained by PCR. The nucleotide sequence information of each primer is shown in Table 4. The gene sequence information corresponding to each genome integration fragment is shown in Tab...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com