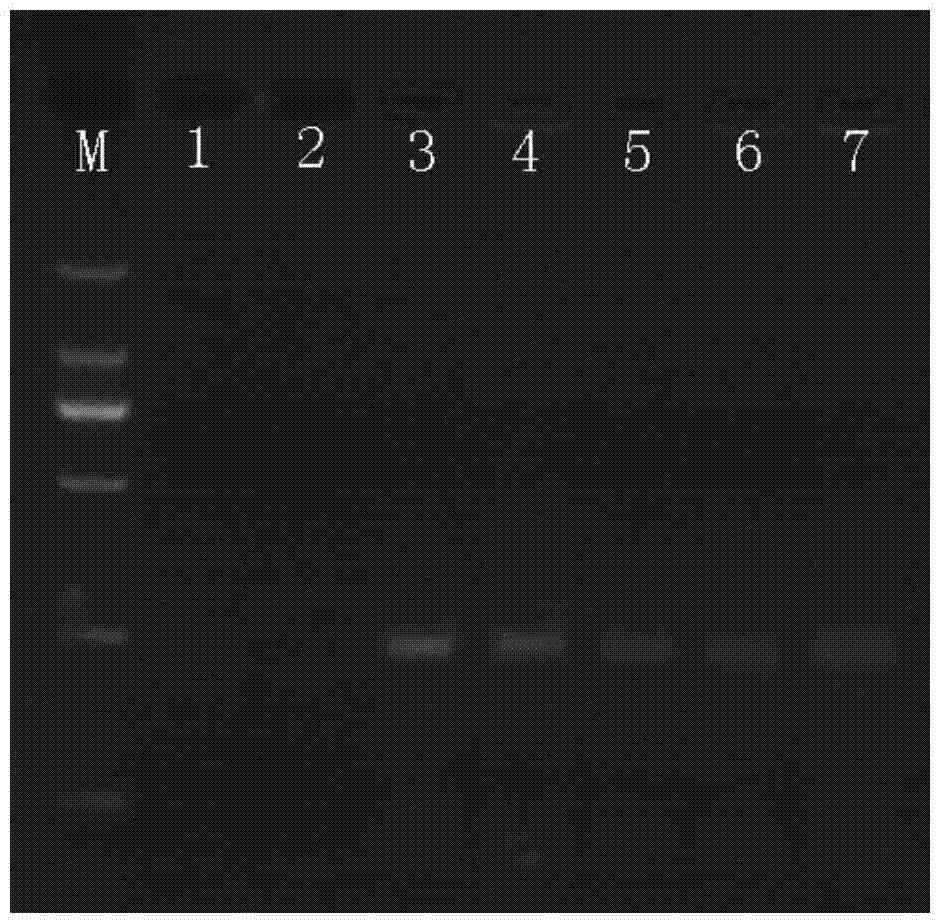Method for detecting pseudomonas syringaepv altinidia through recombinase-mediated isothermal amplification technology
An isothermal amplification, canker bacteria technology, applied in recombinant DNA technology, microorganism-based methods, biochemical equipment and methods, etc., can solve problems such as no application, and achieve high specificity and stability, and the effect of simple and fast reaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 1. Primer design:
[0041] In this experiment, multiple sets of RPA primers were designed according to the 16S-23S rDNA sequence of P. kiwifruit canker sores, and then two sets of primers were selected for RPA amplification reaction. The sequences of these two sets of primers are:
[0042] Primer name
Sequence 5'-3'
RPA Psa1-F
CCTGATAAGGGTGAGGTCGGCAGTTCGAATC
RPA Psa1-R
TCACGCACCCTTCAATCAGGATGGAATGCTC
RPA PSA2-F
AGAAGCAGCTTTTGCTTTGCACACCCGATTTT
RPA PSA2-R
AATCAACATTCACGCACCCTTCAATCAGGATG
[0043] 2. The cultivation of kiwifruit canker bacteria:
[0044] The kiwifruit canker bacteria were put into the liquid LB medium and cultured at 22°C for 12 hours.
[0045] 3. Total DNA extraction of canker bacteria:
[0046] Take 1ml of the above-mentioned cultured bacteria solution, centrifuge to collect ulcer bacteria, extract DNA according to the instructions of the bacterial DNA extraction kit, measure the concentr...
Embodiment 2
[0051] Example 2: Rapid Analysis of RPA
[0052] According to the primers identified in Example 1 above, several reaction time gradients were designed, namely 5min, 10min, 20min, 30min, 40min, 60min, 120min, and the influence of RPA reaction time on detection was analyzed. It was found that the target bands could not be detected after 5 minutes and 10 minutes of reaction, and the target fragments could be detected after 20 minutes, but the amplified fragments did not increase with the prolongation of the reaction time, indicating that RPA has high enzymatic activity and can be detected in a relatively short time. The DNA fragments were amplified within a certain time, and the experiment determined that the optimal reaction time of the RPA reaction was 20 min. Experimental results such as figure 2 shown.
Embodiment 3
[0053] Embodiment 3: Sensitivity analysis of PCR and RPA detection
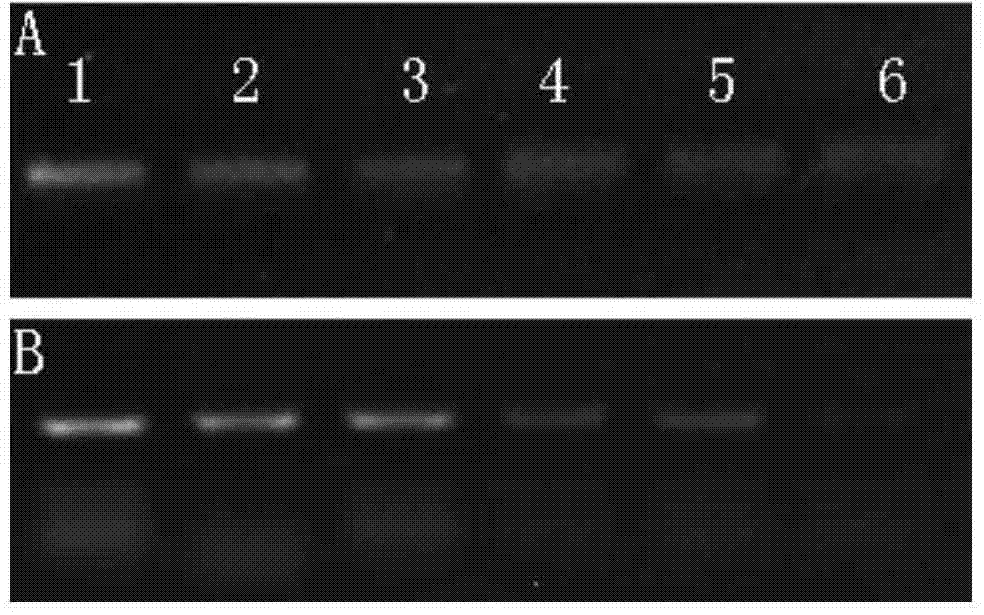
[0054] The total DNA of P. kiwifruit canker was diluted to 50ng / μL, and then diluted according to 50 times, 100 times, 200 times, 300 times and 600 times for PCR and RPA detection. The primers used for PCR detection refer to Rees-George (Rees-George, J., Vanneste, J.L., Cornish, D.A., Pushparajah, I.P.S., Yu, J., Templeton, M.D., and Everett, K.R. 2010. Detection of Pseudomonas syringae pv.actinidiae using polymerase chai n reaction (PCR) primers based on the 16S-23S rDNA intertranscribed spacer region and compari son with PCR primers based on other gene regions.Plant Pathol.59:453-464) published primers, wherein the upstream primer 5'-TTTTGCTTTGCACACCCGATTTT-3'; the downstream primer is 5'-CACGCACCCTTCAATCAGGATG-3'. The experimental results show that both PCR and RPA can see the target fragment when the concentration of 50ng / μL DNA is diluted to 600 times, which proves that RPA and PCR have high detection s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com