Lipase, engineering bacterium and preparing methods of the lipase and the engineering bacterium
A lipase and engineering bacteria technology, applied in the field of genetic engineering, can solve the problems of low enzyme activity and unsatisfactory economic benefits, and achieve the effects of improving unit enzyme activity, good industrial applicability, and improved expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0057] The preparation method of engineering bacterium provided by the invention is characterized in that, comprises the following steps:
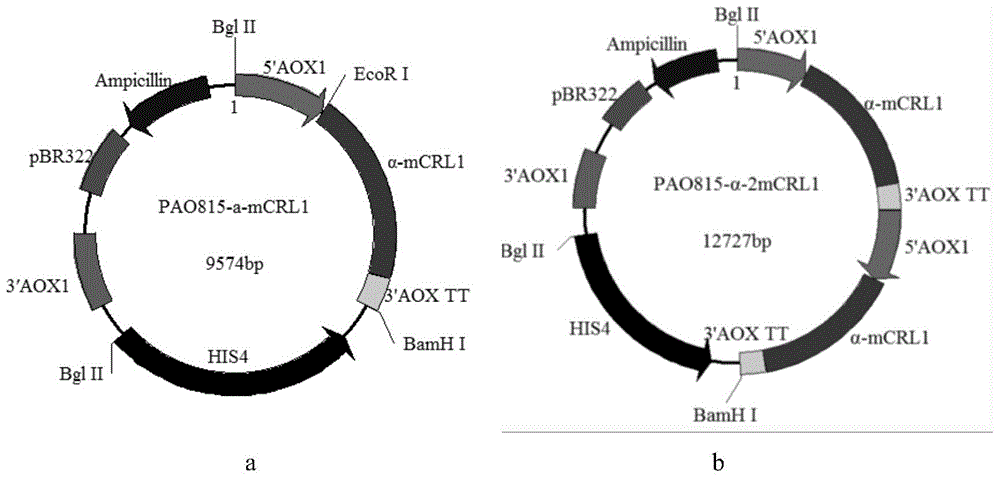
[0058] (1) Construction of lipase gene expression vector: the Pichia pastoris expression system plasmid used for His site insertion is used as a vector, which contains the 5'AOX1 strong promoter, the α-factor signal peptide of Saccharomyces cerevisiae, and the lipase in sequence Gene fragments, the lipase gene fragments include 5'AOX1-TT fragments, His4 (histidine dehydrogenase gene), 3'AOX1, PBR322 and Ampicillin; the Pichia pastoris expression system for His site insertion Plasmid is PAO815 plasmid, its structure is as follows figure 1 shown.
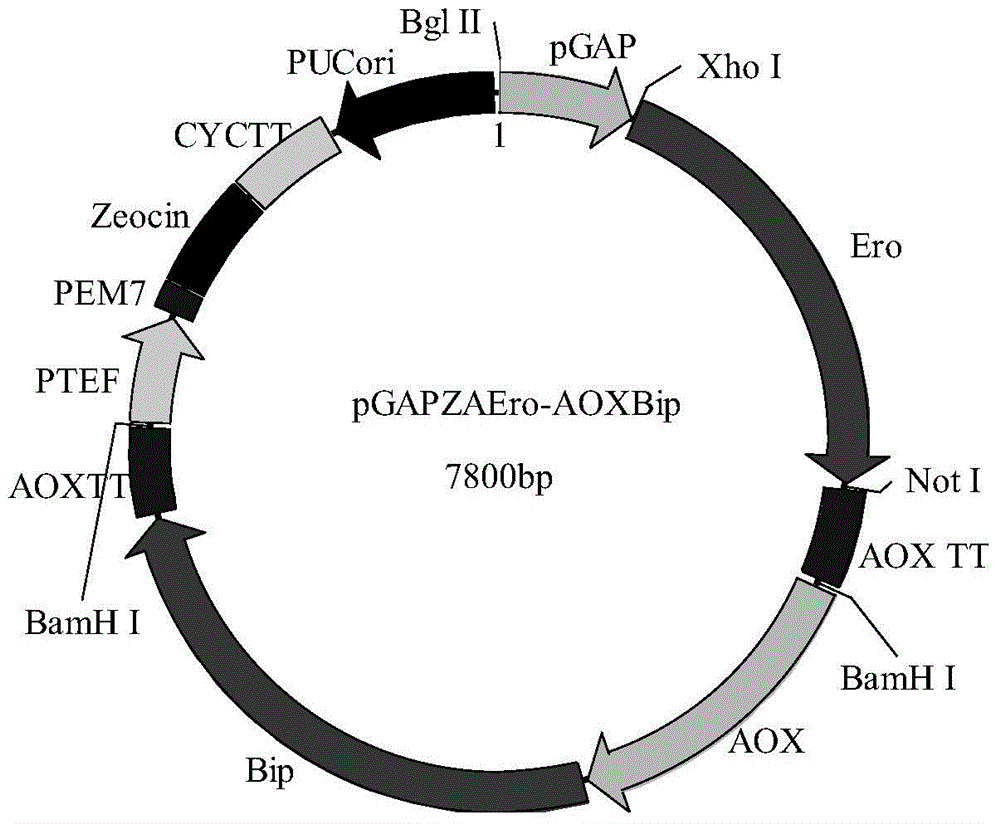
[0059] (2) Constructing a molecular chaperone combination gene expression vector: using the Pichia pastoris expression system plasmid for inserting functional genes at the AOX1 site as a vector, insert two functional genes, wherein the first functional gene contains 5'AOX1 in sequence A strong p...
Embodiment 1
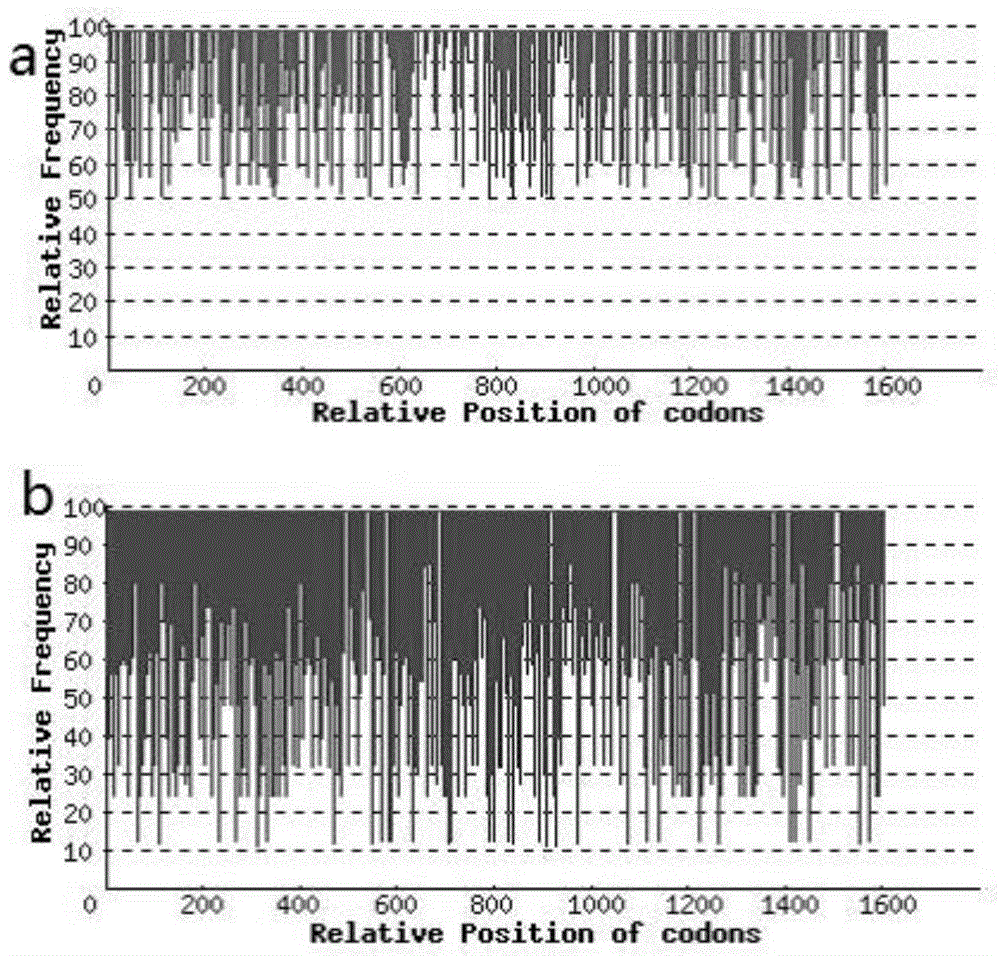
[0073] A lipase whose gene sequence is the gene sequence shown by SEQ ID No.1. Before and after codon optimization, the codon adaptability index changes as image 3 shown. The frequency distribution of codon usage after codon optimization is as follows: Figure 4 shown. GC content in codon usage before and after codon optimization as Figure 5 shown.
Embodiment 2
[0075] A kind of engineering bacterium, its host bacterium is GS115 type Pichia pastoris, gene structure is as follows figure 1 shown.
[0076] At its His site, the gene sequence of the lipase of Example 1 is inserted, and the copy number is 2. The α-factor signal peptide gene sequence of Saccharomyces cerevisiae is inserted immediately upstream of it.
[0077] A molecular chaperone combination gene is inserted at the AOX1 site to increase the expression of the lipase. The molecular chaperone combination gene is pGAPZAEro1p-pPICZABip, and its sequence is the gene sequence shown by SEQ ID No.2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com