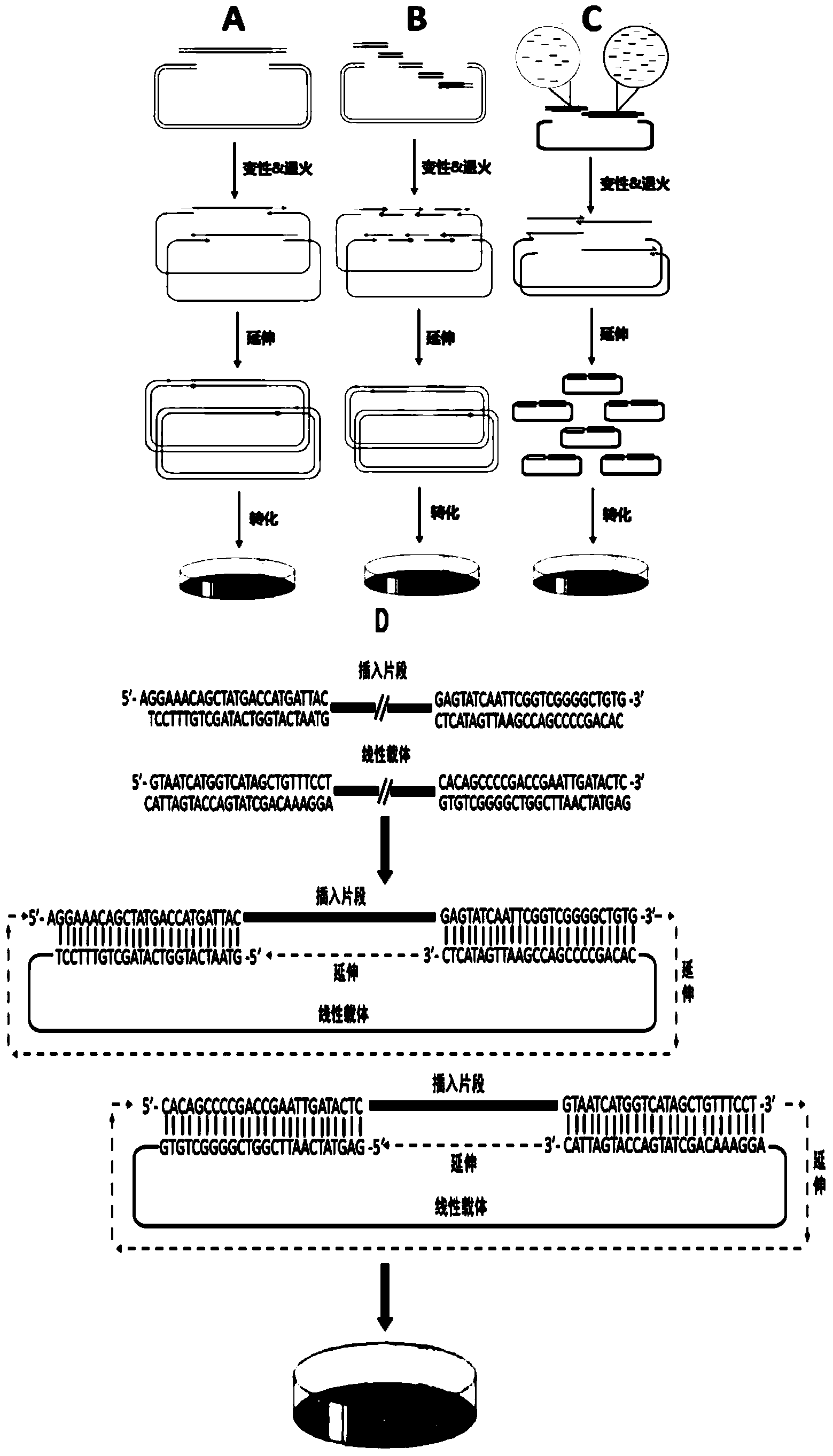
DNA assembly and cloning method
A technology of inserting fragments and linear vectors, applied in the biological field, can solve the problems of no obvious advantages in time and economic cost, increased probability of non-specific hybridization, and increased cloning steps, so as to achieve high reaction efficiency and fidelity. , the effect of low mutation probability and short reaction time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1: Cloning of a single gene.
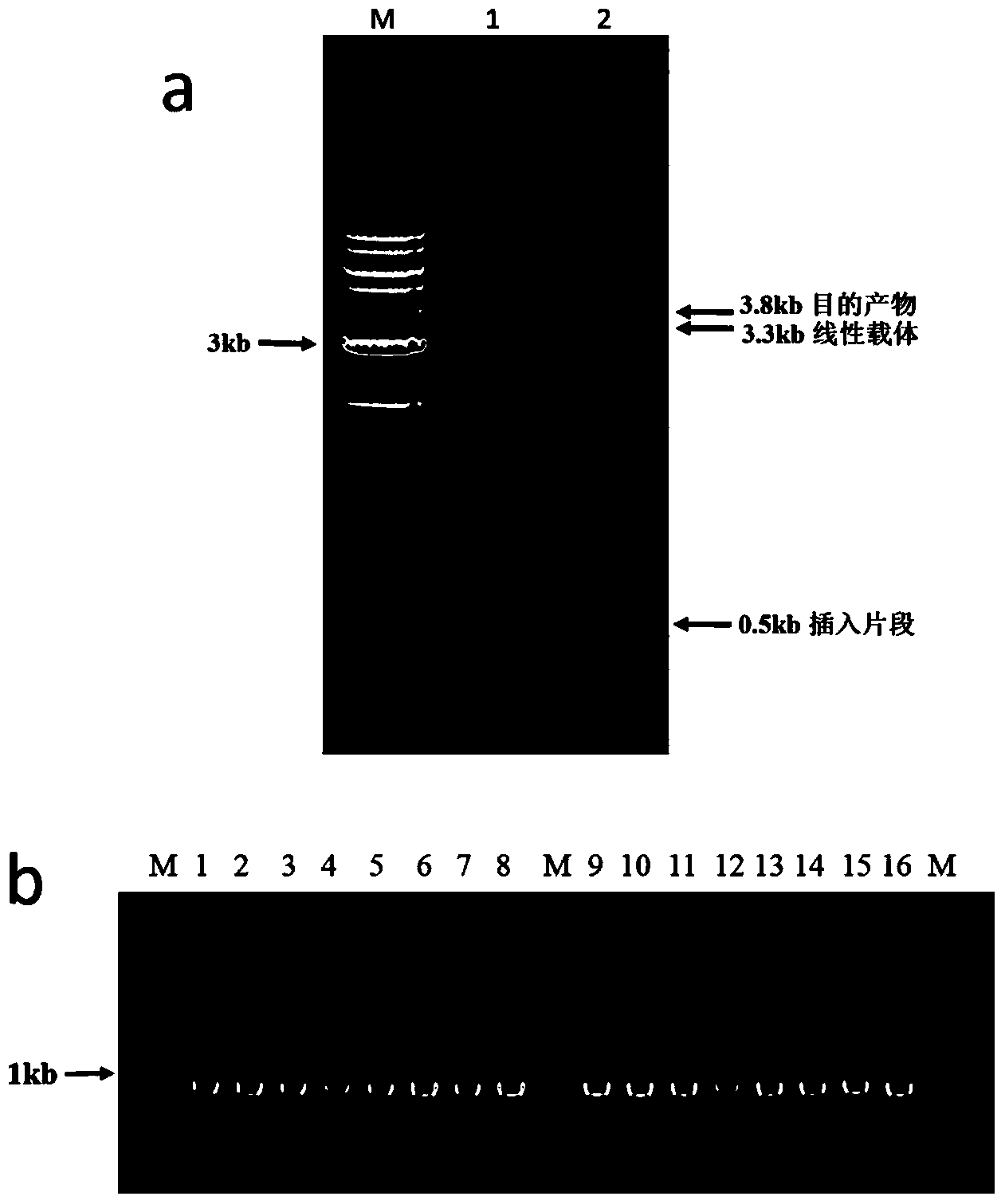
[0081] The pAcGFP1 plasmid (3.3 kb, purchased from clontech) was used as the target vector, and the gentamycin resistance gene (531 bp) was used as the insert fragment for DNA recombination cloning.
[0082] 1. Acquisition of inserts and vectors with overlapping sequences: by respectively designing 2 pairs of corresponding primers (pAcGFP1-F and pAcGFP1-R, Gm-F and Gm-R) to initiate PCR to obtain inserts with overlapping sequence ends and linear carrier. See Table 2 for specific primer sequences.
[0083] The PCR reaction system in which the vector pAcGFP1 is linearized: 2ng pAcGFP1 plasmid as a template, 0.5μM primers (pAcGFP1-F and pAcGFP1-R), 0.8mM dNTP mixture (purchased from NEB, USA), 1×Phusion HF buffer (purchased from NEB Company of the United States), 0.5 μl of Phusion DNA polymerase (purchased from NEB Company of the United States), and replenish water to 50 μl. PCR reaction conditions: 98°C, 30s; (98°C, 10s; 58°C, 30s...
Embodiment 2
[0088] Example 2: Assembly and cloning of multi-gene fragments.
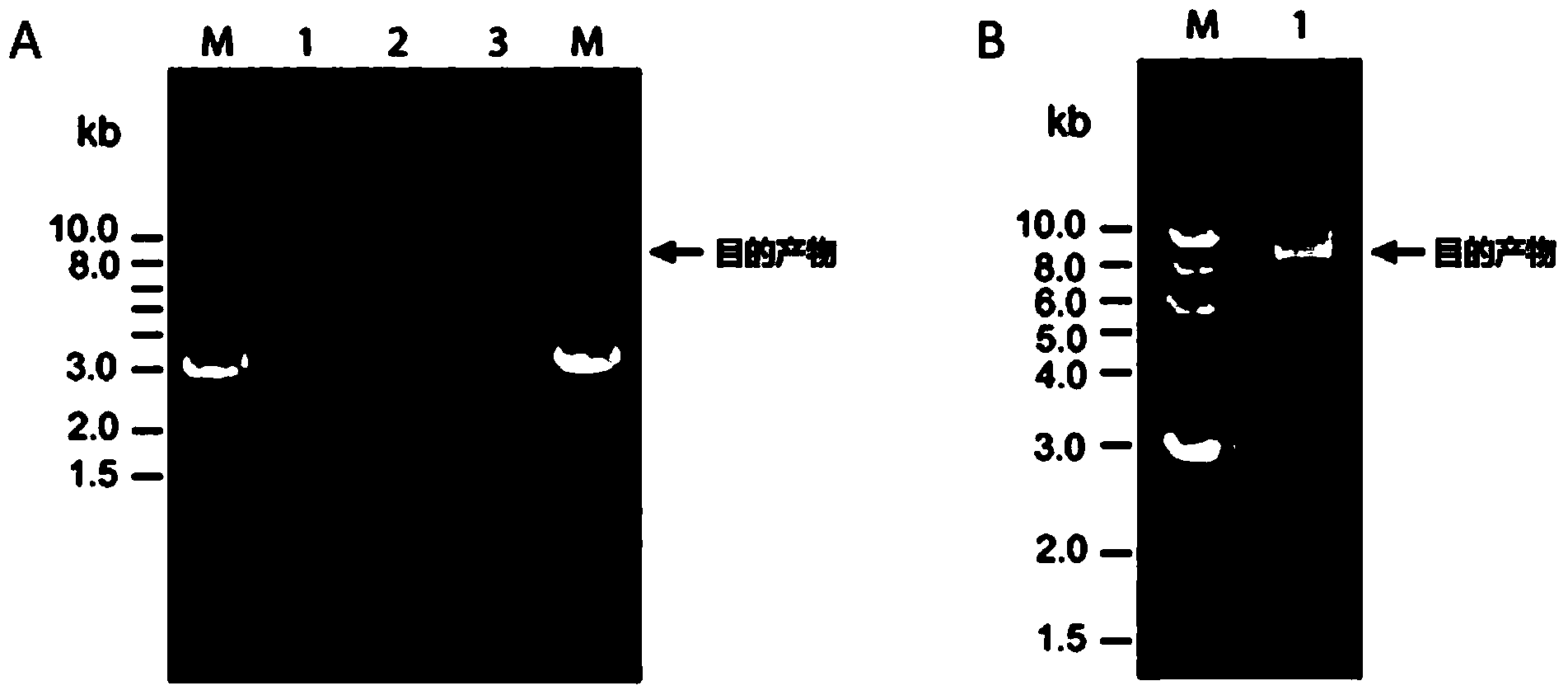
[0089] Take the assembly and cloning of 4 genes (3280bp cat2phaC, 2959bp pASK, 2047bp phaAB, 171bp terminator) in the synthetic pathway of a biodegradable plastic poly(3HB-co-4HB) as an example:
[0090] 1. According to the DNA sequences of the 4 genes, design and synthesize 4 pairs of primers respectively, and use the corresponding plasmids as templates to amplify the 4 genes. See Table 2 for specific primer sequences. Refer to Step 2 in Example 1 for the PCR reaction system and conditions. The PCR products were purified by electrophoresis and recovered.
[0091] 2. Four genes were added to the reaction system in equal molar amounts (3280bp gene 200ng) for assembly, and 4 sets of PCR reactions were performed in parallel, and the cycle numbers were 2, 5, 10 and 20, respectively. For the reaction conditions, refer to step 3 in Example 1. The product electrophoresis results were as image 3 shown. Depend on ...
Embodiment 3
[0093] Example 3: Assembly and cloning of a multi-gene fragment library.
[0094] Taking pAcGFP1N1 (4.7kb) as the target vector, and the gene fragment library (1.7kb) of the HIV envelope gene gp120 synthesized according to codon degeneracy as the insert fragment, DNA assembly and cloning were performed as an example. The specific operations are as follows:
[0095] 1. Chemically synthesize the HIV envelope gene gp120 gene fragment library containing merged bases, and use two methods to obtain recombinant plasmids containing the target gene:
[0096] Method a: Two-step assembly, that is, first assemble the small DNA fragments in the gene fragment library into a full-length gene, and then connect with the carrier;
[0097] According to the DNA sequences of the pAcGFP1N1 and gp120 gene fragment libraries, three pairs of primers were designed and synthesized: pAcGFP1N1-R and pAcGFP1N1-L (for the linearization of the pAcGFP1N1 circular vector), gp120-R and gp120-28L and gp120-L and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com