Application of rice nitrate transporter nrt1.1b in improving plant nitrogen use efficiency
A technology of NRT1.1B, nitrate transport, applied in application, plant peptides, plant products, etc., to achieve the effect of increasing nitrate content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
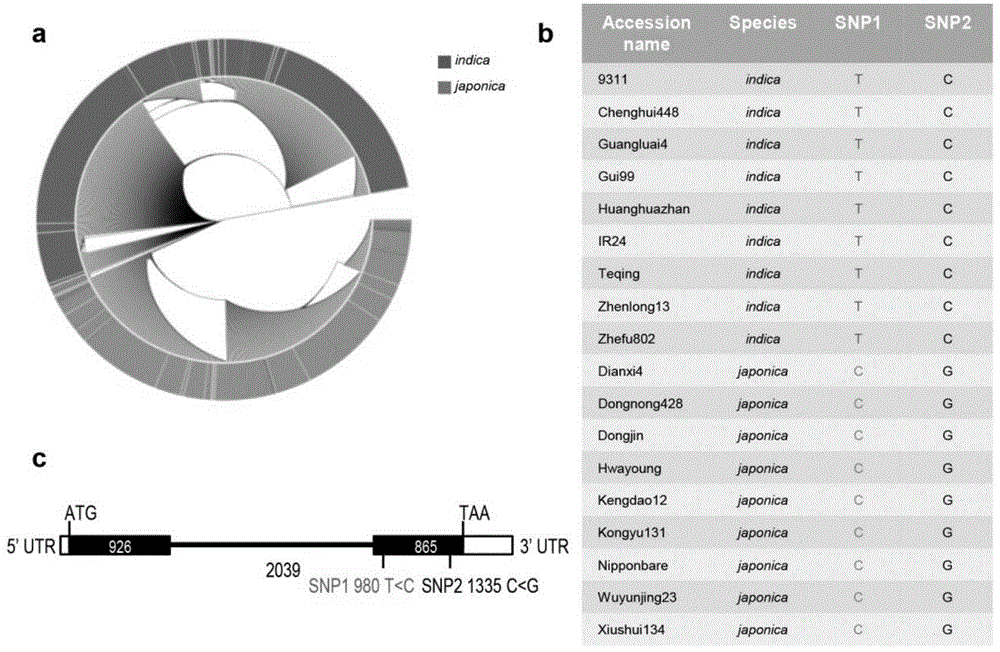
[0035] Example 1, Nucleotide variation in the CDS region of indica and japonica NRT1.1B
[0036] Through the evolutionary analysis of large-scale rice varieties, the results showed that the nitrate transporter gene NRT1.1B (LOC_Os10g40600) showed significant differentiation between indica and japonica varieties ( figure 1 a). The results of sequence analysis showed that there were two single nucleotide variations (SNP1, SNP2) in the CDS region of NRT1.1B in indica and japonica rice, such as figure 1 b and figure 1 As shown in c, and the SNP1 (T<C) at the 980th position in the CDS region leads to amino acid variation (Met<Thr) between indica and japonica rice subspecies.
Embodiment 2
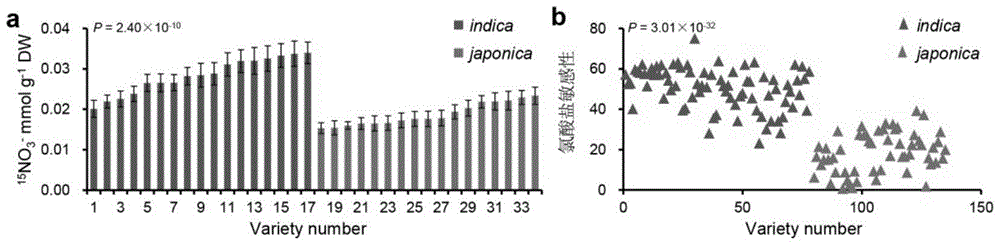
[0037] Embodiment 2, indica rice and japonica rice 15 NO 3 - Content and Chlorate Sensitivity Assays
[0038] The composition of the improved Kimura nutrient solution includes: 2mM KNO 3 , 0.36mM Ca(Cl) 2 4H 2 O, 0.54mMMgSO 4 ·7H 2 O, 0.18mM KH 2 PO 4 , 40μM Fe(II)-EDTA, 18.8μM H 3 BO 3 , 13.4μMnCl 2 4H 2 O, 0.32 μM CuSO 4 ·5H 2 O, 0.3 μM ZnSO 4 4H 2 O and 0.03 μM Na 2 MoO 4 4H 2 O.
[0039] The seedlings of indica and japonica rice were cultured for 2 weeks and then transferred to the seedling containing 5mM 15 N-KNO 3The improved Kimura nutrient solution was absorbed for 24 hours, then treated with 0.1mm CaSO4 for 1 minute, rinsed with deionized water for 3 times, and the aerial part was dried at 70°C. The dried samples were ground to powder, and the 15N content was determined using an isotope ratio mass spectrometer (Thermo Finnigan Delta plus XP; Flash EA1112). The results showed that indica cultivars exhibited higher nitrate content than japonica cu...
Embodiment 3
[0041] Example 3, Determination of Nitrate Transport Ability of Indica and Japonica NRT1.1B in Xenopus Oocytes
[0042] The RNAs of the indica rice IR24 and the japonica rice Nipponbare were extracted and reverse transcribed into cDNA. The cDNAs obtained respectively were used as templates, and the NRT1.1B CDS of indica rice and japonica rice were respectively amplified by PCR using the following primer sequences. The two ends of the primers used for amplification were respectively introduced into the recognition sites of restriction endonucleases BamH I and EcoR I (as shown in the underline), primer sequence: F: 5'- GGATCC ATGGCGATGGTGTTGCCG-3';R:5'- GAATTC TTAGTGGCCGACGGCGATGGT-3'.
[0043] The amplified fragments of interest were ligated into the Xenopus oocyte expression vector pCS2+, and were transcribed in vitro using the mMESSAGEmMACHINE (Ambion; AM1340) kit. Injection and culture of cRNA were performed according to the reported method (Zhang et al., 1998). The cu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com