Aureobasidium pullulans malate-CoA ligase as well as recombinant expression vector and application thereof
A technology of malyl coenzyme and Aureobasidium pullulans, which is applied in the field of genetic engineering, can solve the problems that the sequence has not been reported, the polymalic acid polymerization pathway and related genes have not been elucidated, and achieve the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Embodiment 1, the cloning of malyl-CoA ligase gene
[0025] Using bioinformatics to analyze the reported malyl-CoA ligase gene sequences in NCBI, the sequences were from Phaeospirillum molischianum (WP_002730336); Brucella (Brucell, WP_008503553a); Capsular Rhodobacter (Rhodobacter capsulatus , WP_013066464); Rhodospirillum rubrum (WP_011388966); Paracoccus denitrificans (Paracoccus denitrificans, WP_01174690); after the sequences were compared and analyzed, a demerger primer was designed in the conserved region, and the upstream primer was: 5'-argghggtatggacattgagg- 3' (SEQ ID NO.1), the downstream primer is: 5'-ccrccraaratgttgacraag-3' (SEQ ID NO.2), wherein r=a / g, y=c / t, m=a / c, k= g / t, s=c / g, w=a / t, h=a / c / t, b=c / g / t, v=a / c / g, d=a / g / t, n= a / c / g / t), and then use Aureobasidium pullans CCTCC NO: M2012223 genome as a template for PCR amplification. The amplification conditions are: 94°C pre-denaturation for 5 minutes; Anneal for 30s, extend at 72°C for 30s, 30 cycles; e...
Embodiment 2
[0031] Example 2 Construction of malyl-CoA ligase cDNA recombinant expression vector
[0032] In order to realize expression in Escherichia coli, the total RNA of Aureobasidium pullulans was extracted with a fungal RNA extraction kit (purchased from OMEGA Company, product number R6840-01), and then reverse transcriptase kit (purchased from Fermentas company, product number is K1621) to synthesize the first strand of cDNA, and use this as a template, the upstream primer is: 5'-atgttcaagctcgcccgcag-3' (SEQ ID NO.11), and the downstream primer is: 5'-ttagataccgagagagaactcgacac-3' (SEQ ID NO. 12) Perform PCR amplification. The PCR amplification conditions are: pre-denaturation at 94°C for 5 minutes; denaturation at 94°C for 30 seconds, annealing at 57°C for 50 seconds, extension at 72°C for 2 minutes, and 30 cycles; extension at 72°C for 10 minutes; and finally, incubation at 25°C for 10 minutes. The amplified product was connected to pMD19-T Vector, then transformed into E.coli D...
Embodiment 3
[0033] Example 3 Expression and purification of malyl-CoA ligase
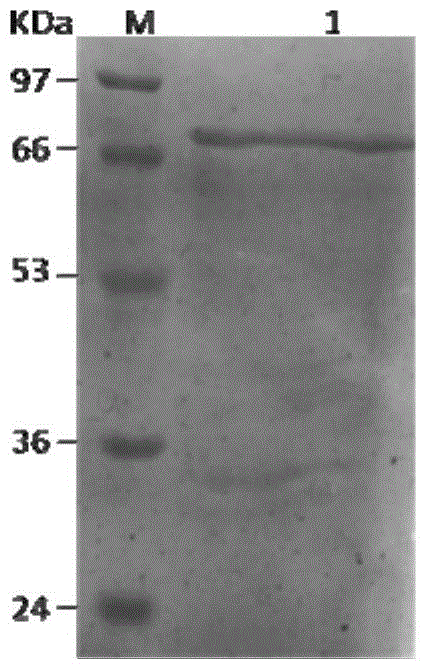
[0034] The constructed recombinant expression vector pET-Mcl was transferred into E.coli BL21(DE3), and after pre-cultivation and 0.5mmol / L IPTG induction for 6h, the bacteria were collected, resuspended in the cell lysate, and used The cells were lysed by sonication, and the cell supernatant was obtained by centrifugation; the protein was purified from the cell supernatant by Ni-NTA affinity chromatography. Then use SDS-PAGE to detect the protein, the results are as follows figure 1 shown. Depend on figure 1 It can be seen that a single band appears at about 68kDa on lane 1. Since the carrier contains four protein tags, Trx.Tag, S.Tag and 2 His.Tag tags, the molecular weight of the purified protein is greater than 48kDa. It indicated that malyl-CoA ligase was successfully expressed, and the protein purification effect was better.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com