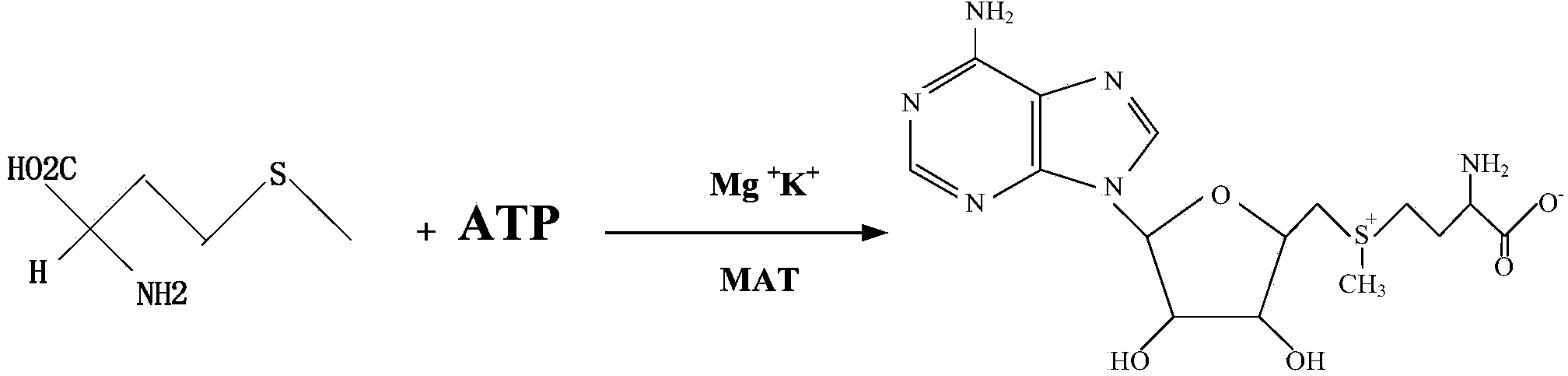
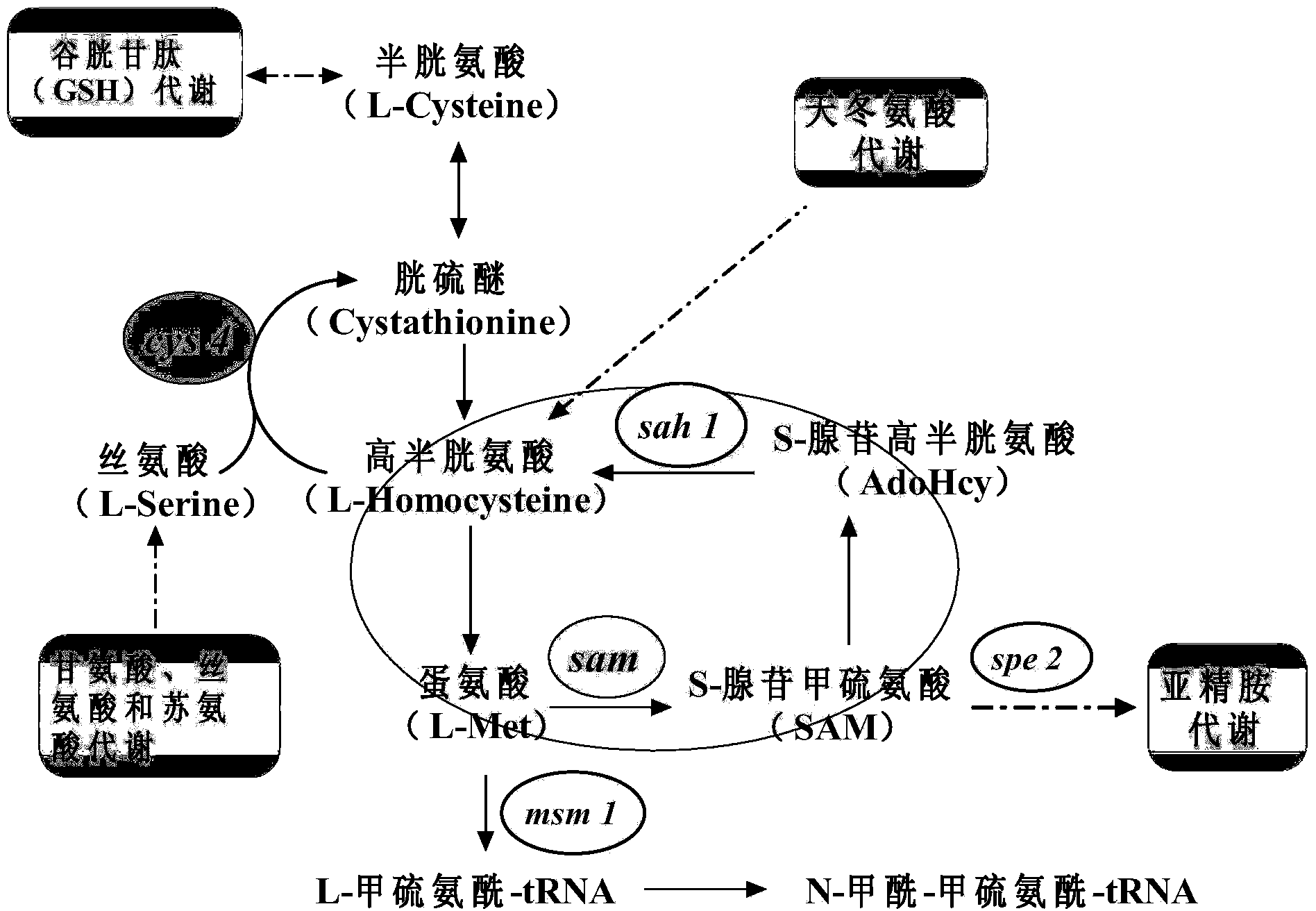
Method for improving yield of S-adenosylmethionine through gene expression regulation
A technology of adenosylmethionine and adenosylmethionine decarboxylase, which is applied in the field of genetic engineering and can solve the problems of nutritional deficiencies of recombinant bacteria and increased fermentation costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0105] Embodiment 1, the construction of recombinant bacteria
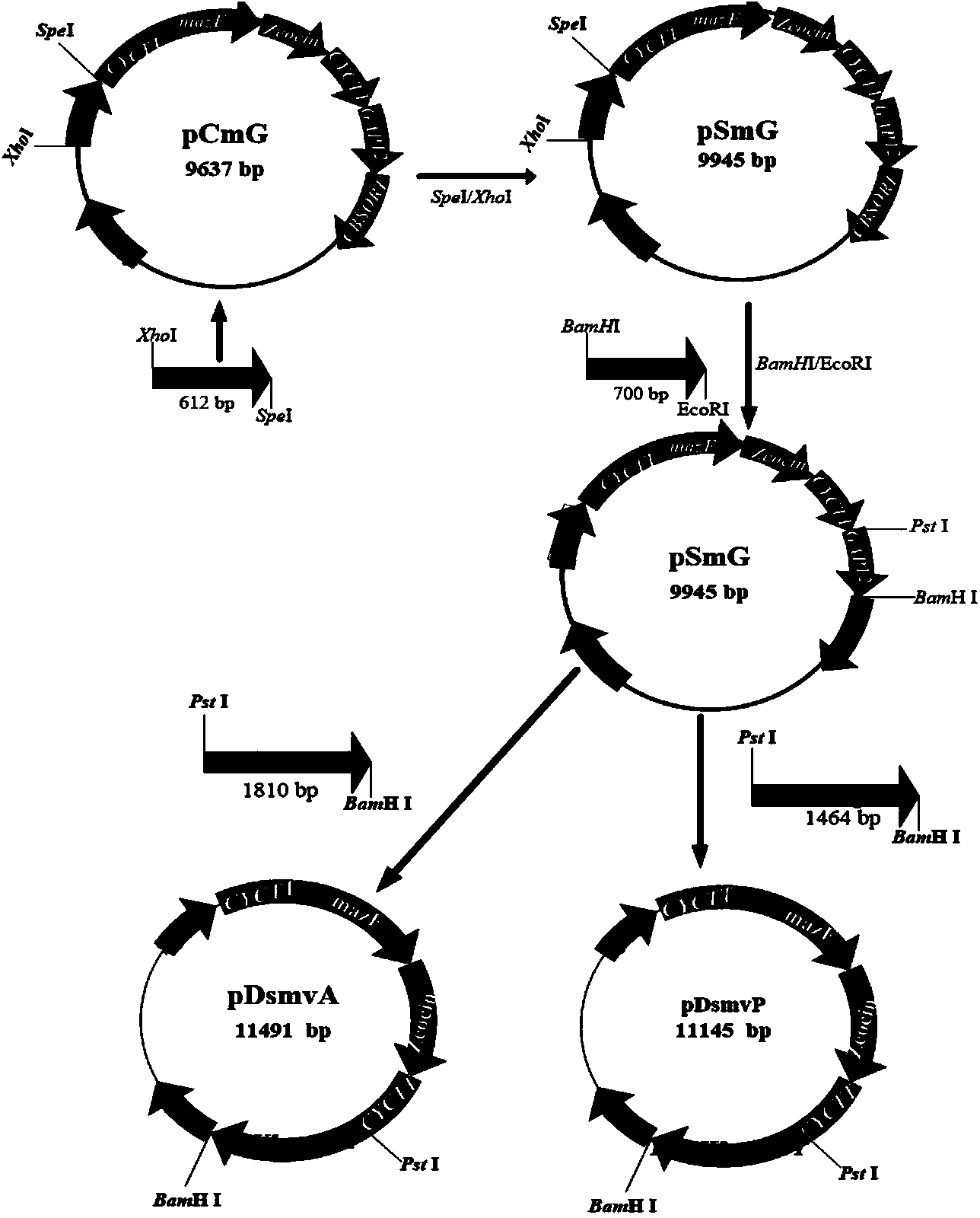
[0106] 1. Construction of pDsmvA and pDsmvP plasmids
[0107] Such as image 3 As shown, using pCmG as the starting plasmid, first replace the homology arm and connect the vgb gene, which are regulated by PADH2 (SEQ ID NO: 3) and PAOX (SEQ ID NO: 2) respectively, wherein PADH2 is a low-lying gene from Pichia stipitis An oxygen-inducible promoter, PAOX is a methanol-inducible promoter in Pichia pastoris, and two plasmids pDsmvA and pDsmvP were constructed. The specific construction process is as follows:
[0108] Using spe2 5'F and spe2 5'R as primers and using Pichia pastoris genomic DNA as a template to amplify Spe2 5' (SEQ ID NO: 4), insert it into the XhoI and SpeI sites of pCmG, replacing the original CBS5; using spe2 3'F and spe2 3'R as primers and using Pichia pastoris genomic DNA as a template to amplify Spe2 3' (SEQ ID NO: 5), insert it into the XhoI and SpeI sites of pCmG, replacing Original CBSORF; A...
Embodiment 2
[0118] Embodiment 2, research on the characteristics of recombinant bacteria
[0119] 1. Shake flask fermentation under normal conditions
[0120] Under normal conditions (250mL shake flask liquid volume is 50mL, rotating speed 220rpm), without taking artificial oxygen limitation measures, culture DS16 / DsvA, DS16 / DsvP and G / Dspe respectively, with DS16 as a control, investigate the effect of vgb expression on the bacteria Effects on growth and survival. The result is as Figure 9 As shown in A, the maximum specific growth rate of DS16 is after induction for 12 hours (0.018h -1 ), the maximum specific growth rate of G / Dspe in 24 hours (0.016h -1 ), and the two strains (DS16 / DsvA, DS16 / DsvP) introduced into the vgb gene, their maximum specific growth rate was 12 hours after induction, and DS16 / DsvA was 0.021h -1 , DS16 / DsvP is 0.023h -1 . It can be seen that after the introduction of the vgb gene, according to the numerical calculation in the figure, the results show that ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com