Identification of antigen-specific adaptive immune responses using Arm-PCR and high-throughput sequencing
A specific and antigenic technology, applied in the direction of immunoglobulin, microbial determination/testing, specific peptides, etc., can solve problems such as time-consuming, hybridoma genetic instability, animal pain and suffering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
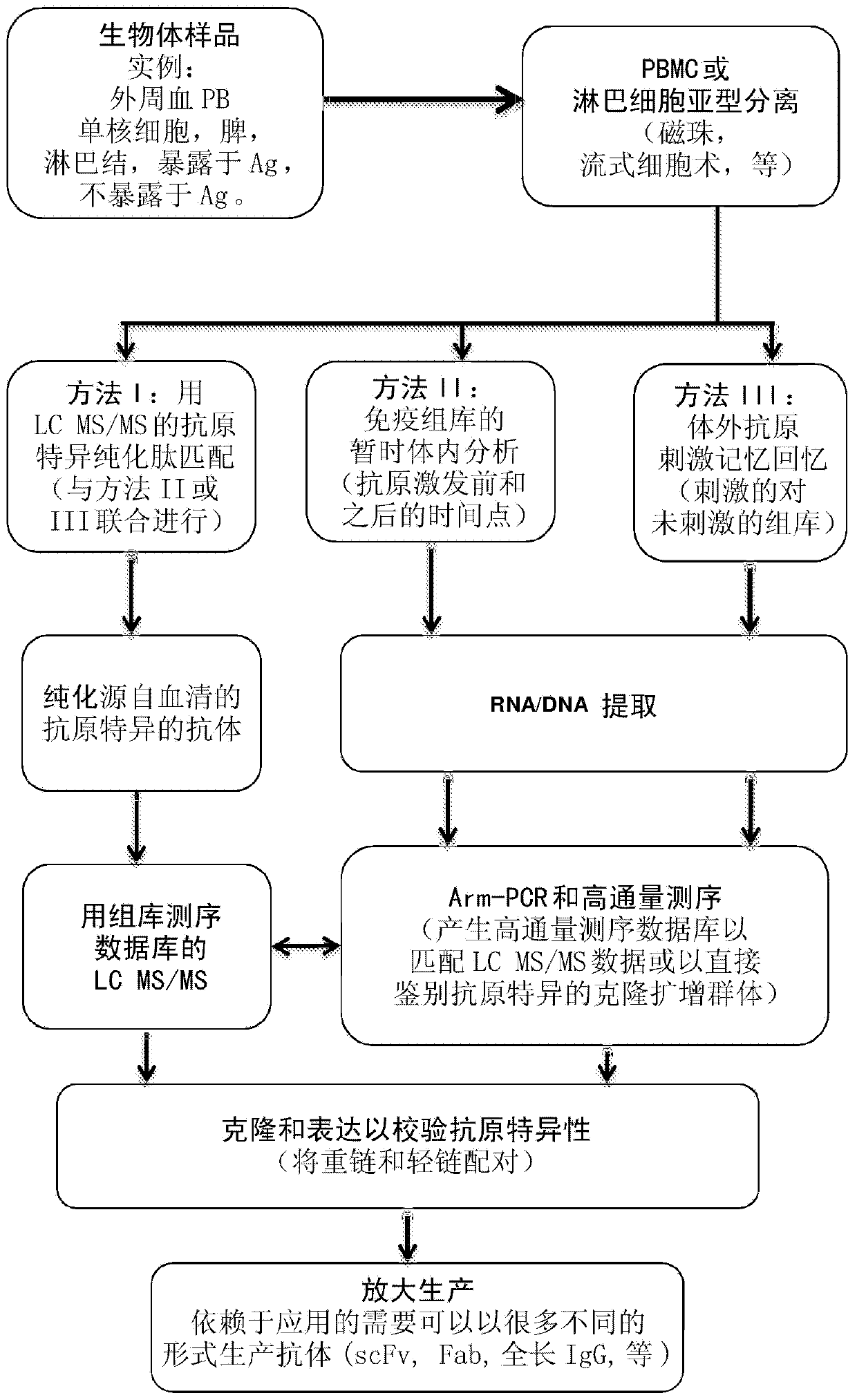
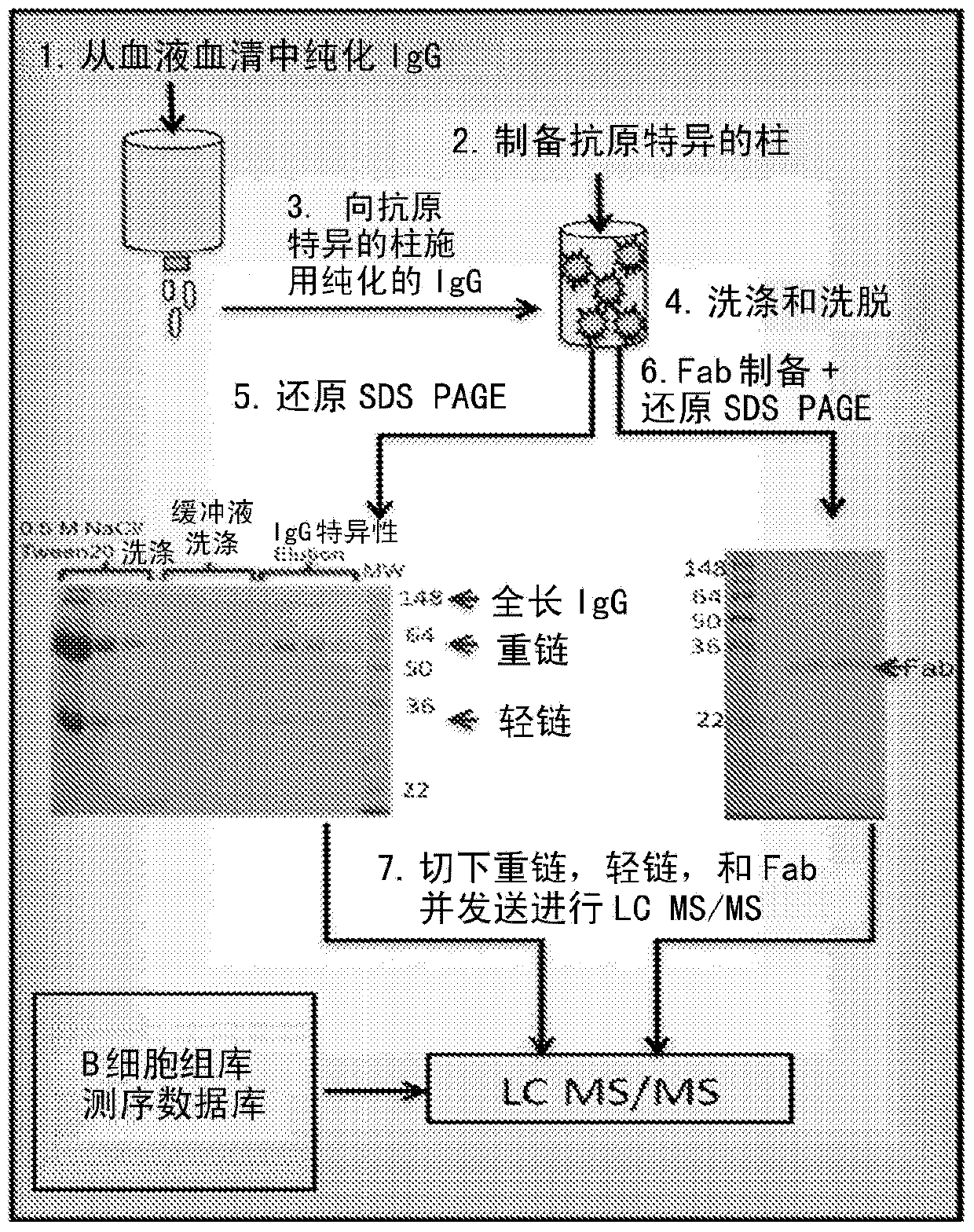
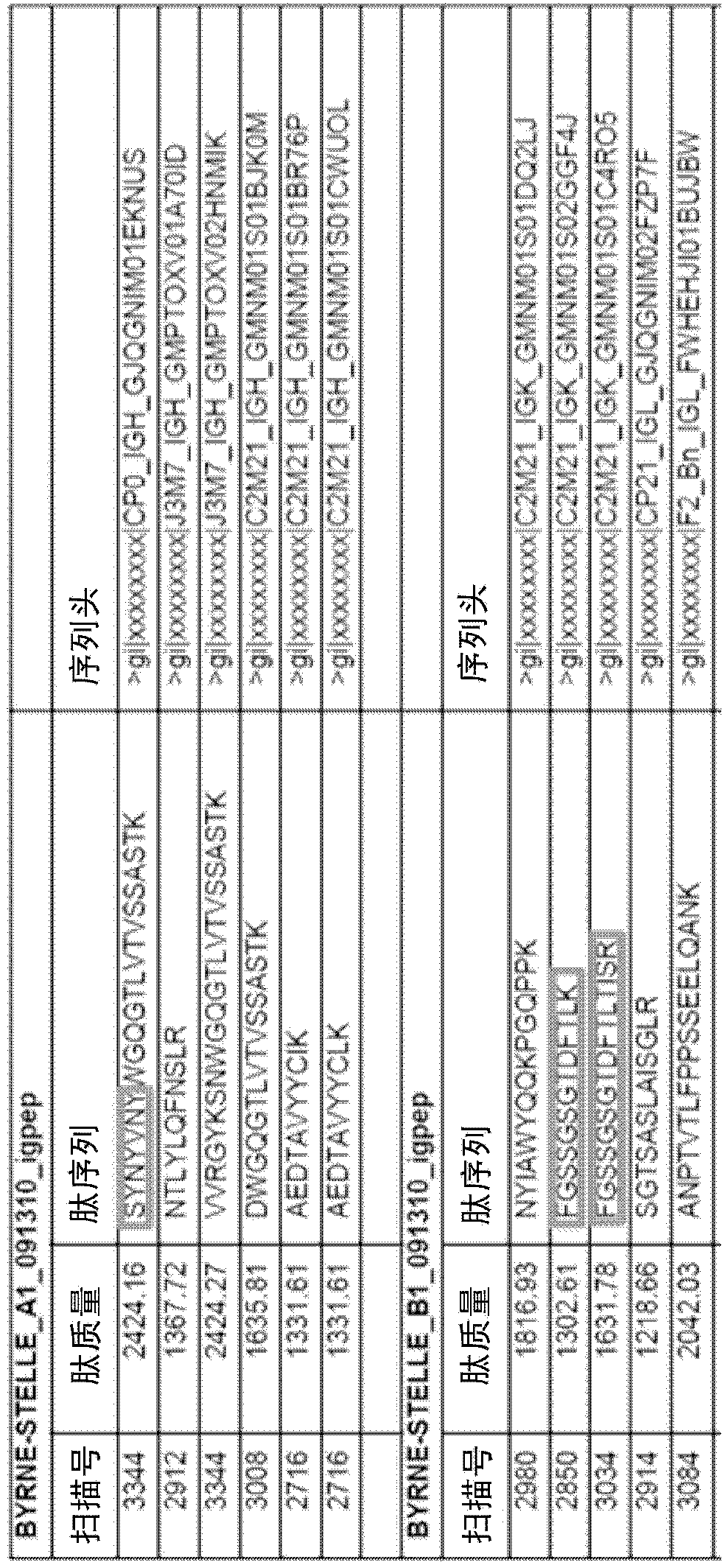
[0039] Hemagglutinin (HA) is an antigenic glycoprotein found on the surface of influenza viruses and used as a component of vaccines to elicit an immune response. The 2009-2010 formulation contained 30 μg / ml HA of each of the following three viruses: A / Brisbane / 59 / 2007, IVR-148 (H1N1), A / Uruguay / 716 / 2007, NYMC X-175C (H3N2) (class A / Brisbane / 10 / 2007 virus), and B / Brisbane / 60 / 2008. Administration of the 2009-2010 influenza vaccine to two healthy volunteers The volunteers reported feeling normal and well for a period of 30 days prior to vaccination. Both the 2008-2009 and 2009-2010 vaccines contained essentially the same influenza A H1N1 and H3N2 antigens but different B-strain antigens. To test our approach and provide the highest probability of matching antigen-specific antibodies to body fluid repertoire sequencing results, we selected volunteers who had previously received the 2008-2009 influenza vaccine.
[0040] Sample Preparation
[0041] Blood samples were taken at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com