Improved sulfite sequencing method
A sulfite and sequencing technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problem of inability to understand the methylation position and state information, the inability to reflect the methylation state of CpG islands, and the purity of DNA Higher requirements and other issues, to achieve the effect of shortening the modification time, shortening the modification time, increasing stability and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] 1. Add 10 μL of EcoR I enzyme, 10 μL of Hand III enzyme and 20 μL of buffer solution to about 10 μg of genomic DNA, add sterile water to 200 μL, and place in a water bath at 37°C for 24 hours for enzyme digestion.
[0021] Wherein the buffer is 100mM Tris-Hcl (pH7.5), 100mM MgCl 2 , 10mM Dithiothreitol, 500mM NaCl;
[0022] 2. After the water bath is over, add chloroform and isoamyl alcohol mixture with an equal volume of 200ul to the enzymatic digestion system, mix intermittently for 10 minutes; then centrifuge at 12000r / min for 10min at 4°C, transfer 180μL of the supernatant to a new 1.5 ml centrifuge tube.
[0023] Then add 1 / 10 volume of 3M NaAC (about 20ul), 40μg glycogen and 3.5 times the volume of supernatant (650μL-700ul) of pre-cooled absolute ethanol, mix well, and precipitate 2 at -20°C. - 3 hours (or overnight); then centrifuge at 4°C, 12000r / min for 30min.
[0024] Discard the supernatant, add 1ml of pre-cooled absolute ethanol to the pellet, mix intermi...
Embodiment 2
[0042] 1. Extract genomic DNA:
[0043] First with 0.1% HgCl 2 The solution was used to sterilize the surface of the selected plump Arabidopsis seeds for 10 minutes, and then wash them with deionized water for 3 times, each time for 5 minutes. After washing, the seeds were soaked in deionized water and placed at 4°C for 3 days to break the dormancy of the seeds. Put the same number of Arabidopsis seeds (about 20 to 30 grains) in a sterilized Erlenmeyer flask, add 100mL containing 0, 0.5 and 5.0mg·L -1 Cd (CdCl 2 Prepared) MS nutrient solution (Murashige and Skoog Basal Salt Mixture), sealed. Place it in a shaker for light culture (temperature 19-20°C, light-dark cycle 14 / 10h, light intensity 3000lx), take samples after 21 days, separate the leaves and roots of Arabidopsis seedlings with razor blades, and separate them with distilled water and sterile water. After washing 3 times, wipe dry with filter paper, and refrigerate in -80°C refrigerator until use.
[0044] Then th...
Embodiment 3
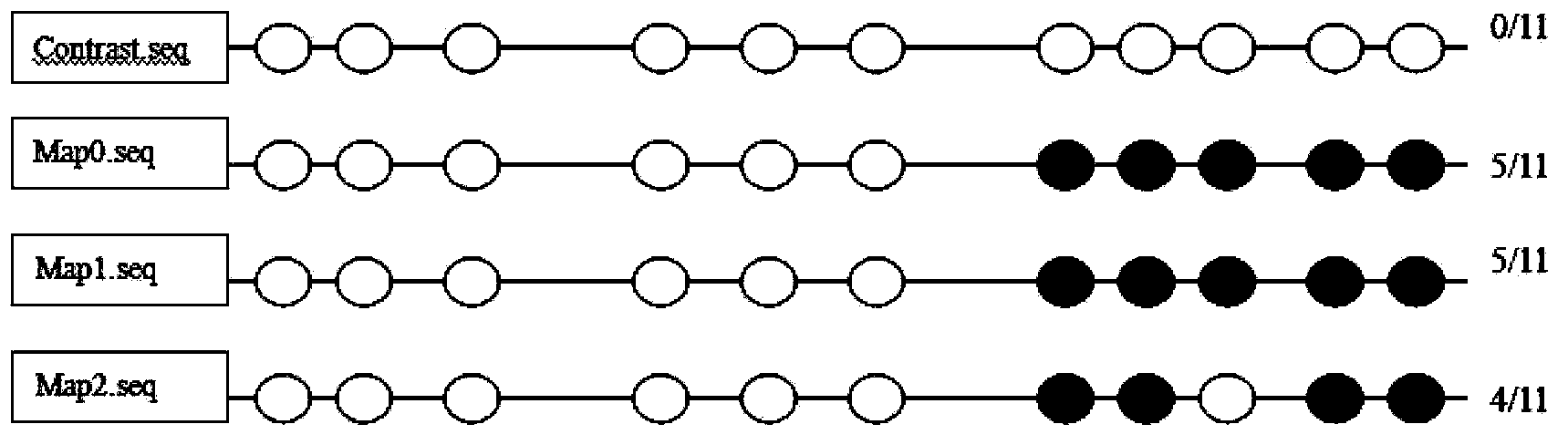
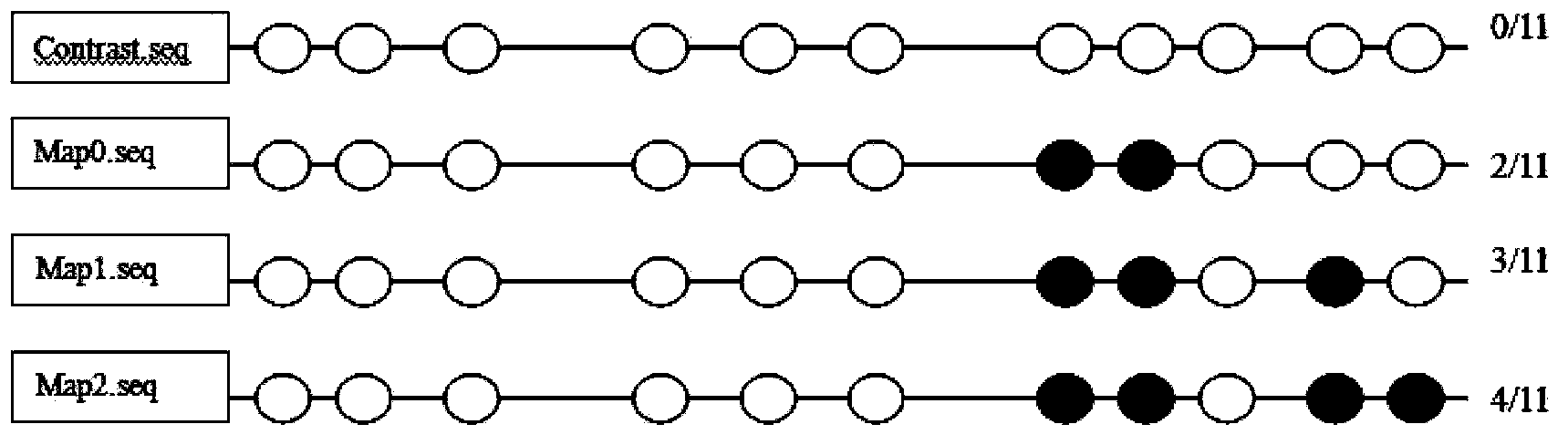
[0077] At the same time, the existing original sulfite sequencing was used for the same sequencing Marianne Frommer (1992), A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands, Proc.Nati.Acad.Sci.USA 89,1827 -1831.), (see figure 1 )
[0078] Depend on figure 1 Compared with the comparison standard, it can be seen that under the stress of Cd concentrations of 0, 0.5, and 5.0, the methylation sites are 5, 5, and 4, respectively. As shown in the figure (a total of 11 CG sites, where the black circle indicates methylation at this site) there is no obvious trend of change.
[0079]
[0080]
[0081]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com