Quick detection method for scallop pathogenic vibrio splendidus
A technology of Vibrio brilliant and detection method, which is applied in the field of aquatic organisms, can solve problems such as being susceptible to interference, unstable results, and low sensitivity of enzyme-linked immunosorbent assay, achieving the effect of ensuring accuracy, reliability and wide detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0027] Experimental Example 1: Specificity Analysis of Primers for Quantitative Detection of Vibrio splendidus:
[0028] The metalloprotease gene in Vibrio candidiasis can be specifically detected by using the primers for quantitative detection of Vibrio candidiasis.
[0029] Vibrio resplendent, Vibrio anguillarum, Vibrio parahaemolyticus, Vibrio alginolyticus, Aeromonas hydrophila, Pseudomonas aeruginosa and Pseudomonas putida were cultured to extract whole genome DNA as PCR reaction template. The general PCR reaction was carried out by using the primers for the quantitative detection of metalloprotease of Vibrio resplendent and the above-mentioned template. The reaction conditions were pre-denaturation at 95°C for 5 minutes; denaturation at 95°C for 30 seconds, renaturation at 60°C for 30 seconds, extension at 72°C for 30 seconds, 30 cycles; extension at 72°C for 10 minutes. After the PCR reaction, the products were detected on agarose gel. The results showed that there w...
experiment example 2
[0030] Experimental Example 2: Sensitivity Analysis of Primers for Quantitative Detection of Vibrio splendidus:
[0031] Vibrio cantilever can be detected with high sensitivity by using the primer for quantitative detection of Vibrio candidiasis metalloproteinase.
[0032] Culture Vibrio splendidus with 2216E liquid culture based on shaking at 18°C for 12 hours, and use the method of plate counting to determine the concentration of the bacterial liquid to be 4×10 9 CFU / ml, use sterile normal saline to dilute the bacterial solution of Vibrio splendidus in a multiple of 10, and the concentrations are 4×10 8 CFU / ml, 4×10 7 CFU / ml, 4×10 6 CFU / ml, 4×10 5 CFU / ml, 4×10 4 CFU / ml, 4×10 3 CFU / ml, 4×10 2 CFU / ml, 4×10 1 CFU / ml, 4CFU / ml. Use the above dilutions of bacteria as templates for common PCR reactions. The reaction conditions are 95°C pre-denaturation for 5 minutes; 95°C denaturation for 30 seconds, 60°C refolding for 30 seconds, 72°C extension for 30 seconds, 30 cycles;...
Embodiment 3
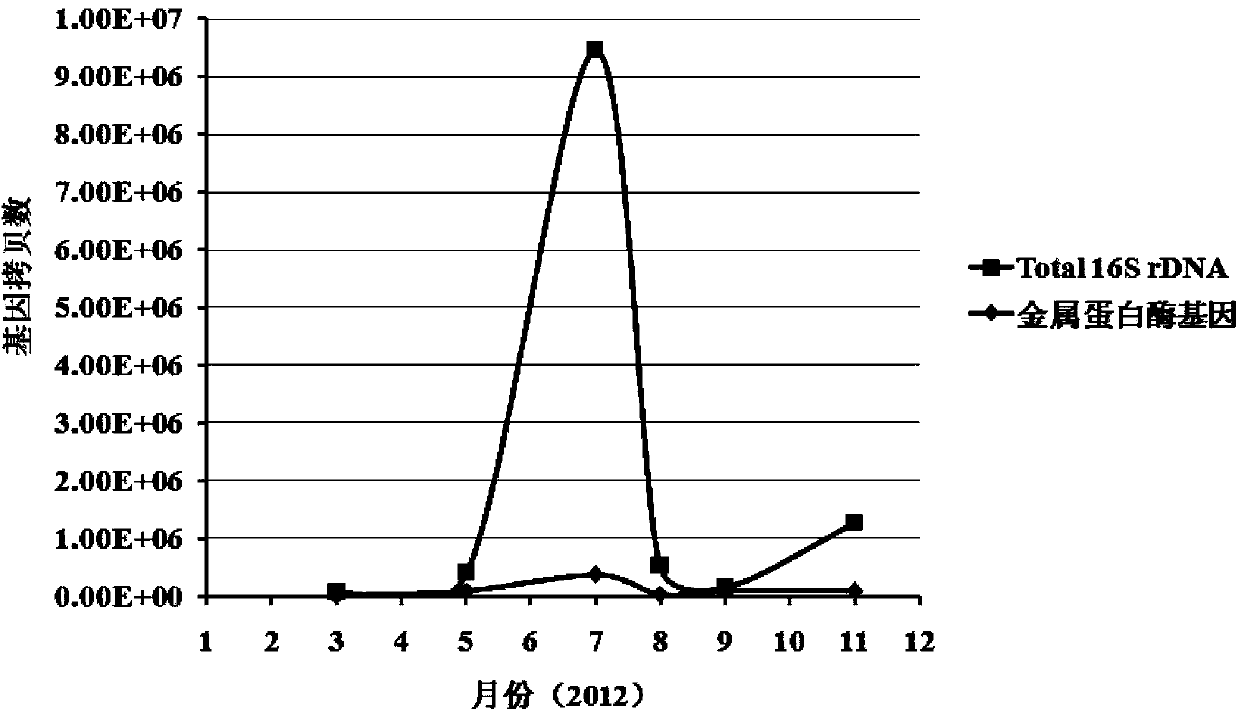
[0034] Example 3: Detection of the proportion of Vibrio splendidus in the culture environment of Dalian scallops
[0035] Water samples were collected from the Dalian scallop culture environment in March, May, July, August, September, and November respectively. The water samples passed through a 0.22 μm filter membrane to enrich the microorganisms in the environment, and the total DNA on the filter membrane was extracted. , to determine the concentration of DNA in environmental samples in each month.
[0036] Before the fluorescent quantitative PCR reaction, the DNA concentration of the environmental samples in each month was diluted to 1ng / μl, and the diluted samples were used for fluorescent quantitative PCR with 16SrDNA and metalloprotease gene primers respectively, and the reaction conditions were pre-denaturation at 95°C for 30 seconds; Denaturation at 95°C for 5 seconds, annealing / extension at 60°C for 30 seconds, collection of SYBR Green fluorescence, 40 cycles. Accord...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com