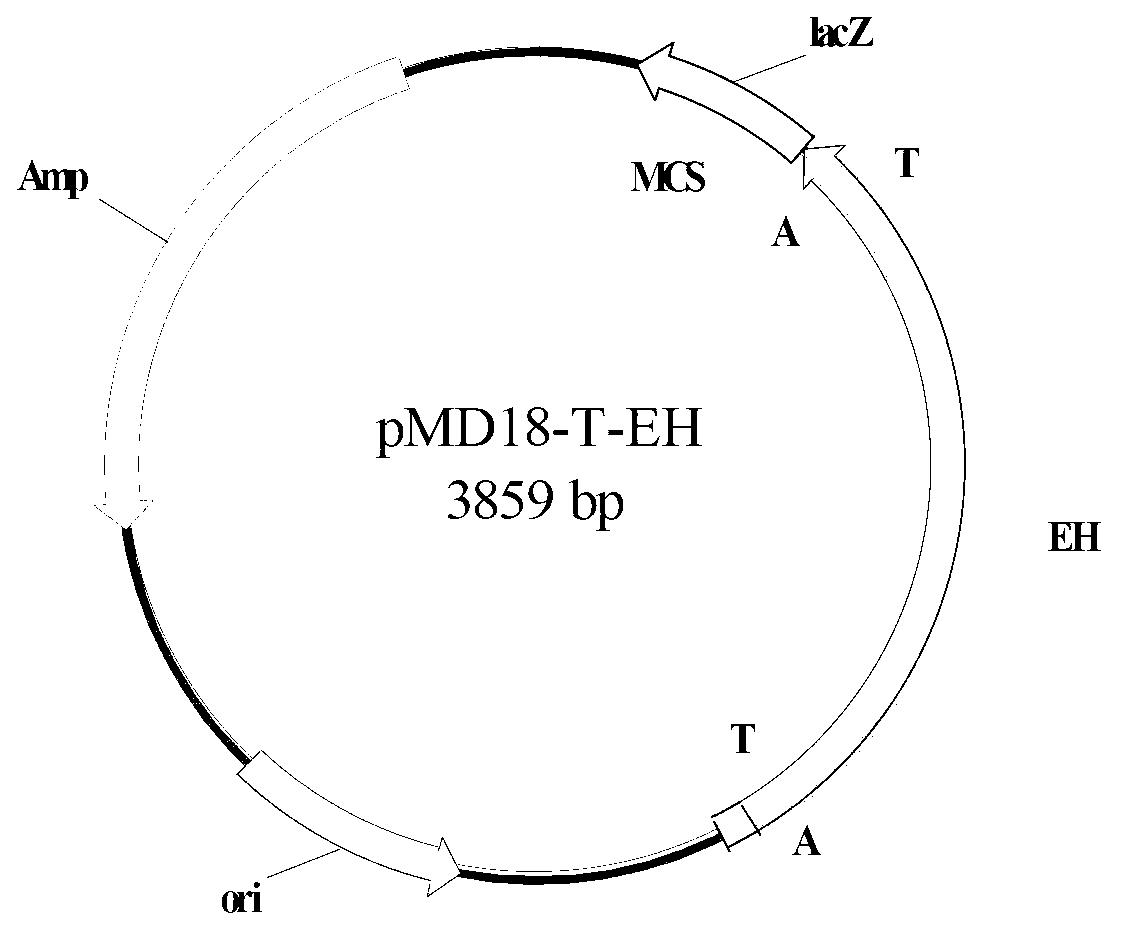
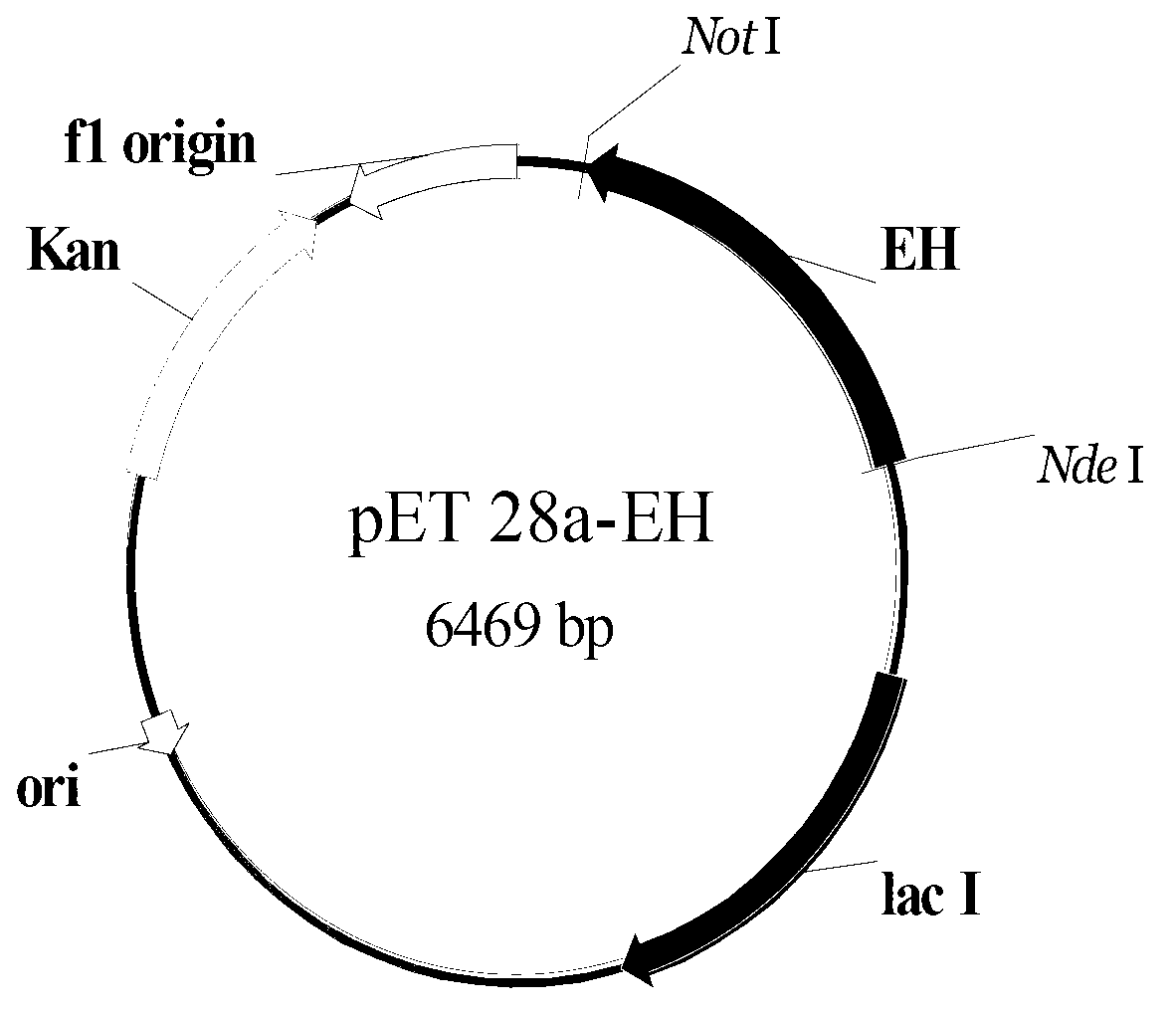
Epoxide hydrolase enzyme gene and encoding enzymeand and carrier and engineering bacteria and application
A technology of genetically engineered bacteria and epoxides, used in genetic engineering, hydrolase, plant genetic improvement, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Acquisition of Agromyces sp. ZJB120203
[0046] The present invention takes soil samples from Zhejiang, Jiangsu, Anhui and other places, and takes 40 parts of soil samples altogether. The specific method of screening: Weigh 5 g of soil samples, add 50 mL of sterile water to make soil suspension, take 1.0 mL of supernatant, add it to the screening medium, shake and cultivate at 30 °C and 150 rpm / min for 4 days. Afterwards, the culture solution was taken for gradient dilution, and 10 -4 、10 -5 、10 -6 Gradient dilutions were spread on the solid medium and cultured in a constant temperature incubator at 30°C for 3 days. The grown single colony was picked and inoculated into the liquid fermentation medium for cultivation. Cultured on a shaker at 30°C for 2 days, the cells were collected by centrifugation, and washed with phosphate buffered saline. The cells are used for transformation, and the transformation conditions are as follows: Add cells to phosphate bu...
Embodiment 2
[0083] Example 2: Cloning of the epoxide hydrolase gene
[0084] The total genomic DNA of Agromyces sp. ZJB120203 cells was extracted with a rapid nucleic acid extraction instrument, and the genomic DNA was used as a template for PCR amplification under the action of primer 1 (YKSCAYGGYTGGCCMGG) and primer 2 (SARYGCRGCAAAGTGTCC). The amount of each component in the system (total volume 100 μL): 10×Taq DNA Polymerase Buffer 10 μL (Mg 2+ ), 0.5 μL of 10 mM dNTP mixture (2.5 mM each of dATP, dCTP, dGTP, and dTTP), 0.5 μL of each cloning primer 1 and primer 2 at a concentration of 50 μM, 1 μL of genomic DNA, 1 μL of Taq DNA polymerase, and 86.5 μL of nucleic acid-free water.
[0085] Using Biorad’s PCR instrument, the PCR reaction conditions were: pre-denaturation at 94°C for 3 minutes, then entering into a temperature cycle of 94°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min, a total of 30 cycles, and finally extending at 72°C for 10 min, with a termination temperature of 8 ℃....
Embodiment 3
[0087] Embodiment 3: Obtaining of epoxide hydrolase gene
[0088] The total genomic DNA of Soil mold ZJB120203 was extracted with a rapid nucleic acid extractor, and PCR amplification was performed under the action of primer 3 (ATGACCGCGGTGAGTCCCAC) and primer 4 (TCATTGTTCATTTCCTCTCCAATTGG) using the genomic DNA as a template. The amount of each component in the PCR reaction system (total volume 100 μL): 10 μL of 10×Pfu DNA Polymerase Buffer, 0.5 μL of 10 mM dNTP mixture (2.5 mM each of dATP, dCTP, dGTP, and dTTP), cloning primer 3 and primer 4 at a concentration of 50 μM 0.5 μL each, 1 μL of genomic DNA, 1 μL of Pfu DNA Polymerase, and 86.5 μL of nucleic acid-free water.
[0089] Using Biorad’s PCR instrument, the PCR reaction conditions were: pre-denaturation at 94°C for 3 minutes, then entering into a temperature cycle of 94°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min, a total of 30 cycles, and finally extending at 72°C for 10 min, with a termination temperature of 8 ℃...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com