Recombinant yeast strain capable of producing conjugated linoleic acid and application thereof
A technology of conjugated linoleic acid and yeast, which is applied in the field of microbial production of conjugated linoleic acid, and can solve the problems of inability to use and low yield of conjugated linoleic acid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] The codon optimization of embodiment 1 linoleic acid isomerase (PAI) gene
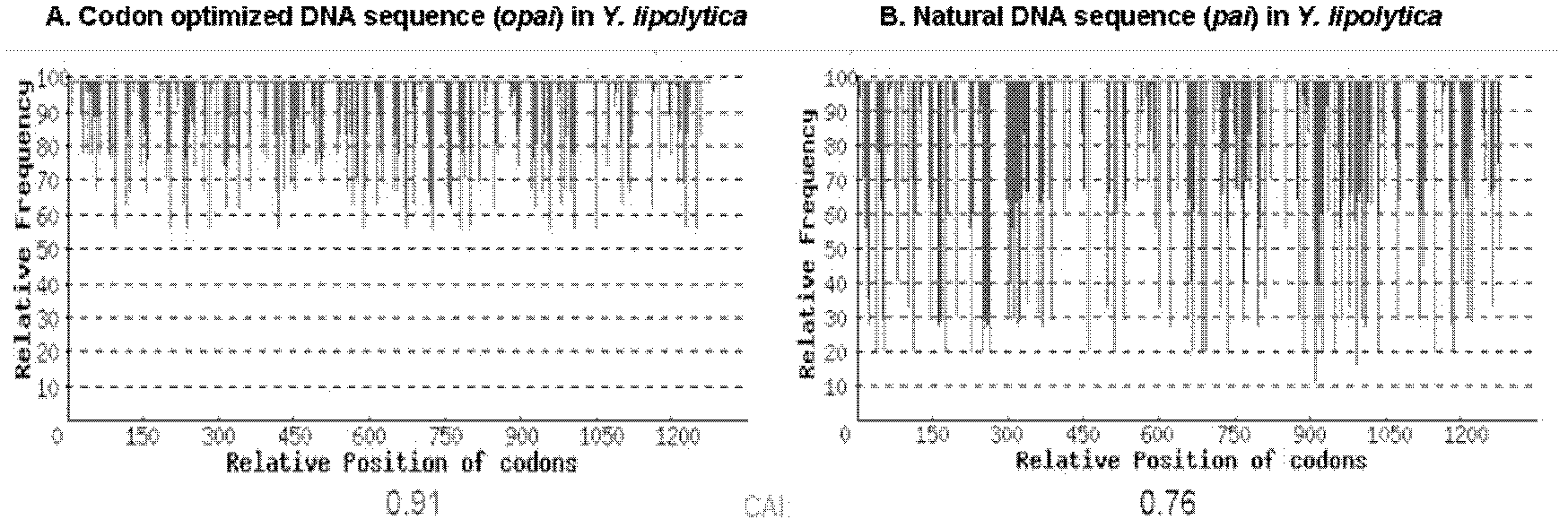
[0020] Referring to the pai gene sequence (genebank: AX062088), based on the characteristics of the Yarrowia lipolytica genome, the algorithm of the Genscript OptimumGeneTM system was used to optimize the design of pai transcription, translation, protein folding and other related parameters, including GC content, rare Codon usage, mRNA structure, and various cis elements during transcription and translation. At the same time, before the start codon of the gene sequence, a Kozak sequence that is conducive to the initiation of translation in eukaryotes: GCCACA is added. The opai nucleotide sequence after codon optimization is shown in SEQID NO: 1, and its codon adaptation index (CAI) has increased from 0.76 to 0.91. For specific results, see figure 1 . The optimized opai gene was synthesized by Nanjing GenScript Biotechnology Co., Ltd., and subcloned into the vector pUC57 (purchased from Nanjing...
Embodiment 2
[0021] Embodiment 2 Construction of recombinant expression plasmid
[0022] Plasmid pGEX6-1-PAI containing the natural linoleic acid isomerase gene pai (genebank: AX062088) derived from Propionibacterium acnes (Prof.Ivo Feussner, Georg-August-University, ttingen, Germany) as a template, with a pair of primers:
[0023] P1: 5′-CACGTGATGTCCATCTCGAAGG-3′ and
[0024] P2: 5′-GGTACCTTACACGAAGAACCGC-3′
[0025] The linoleic acid isomerase gene pai was amplified by PCR. The PCR program is: 15 seconds at 94°C, 30 seconds at 57°C, 1 minute at 68°C, 30 cycles. PCR reaction system: 5 μl 2mM dNTPs, 5 μl 10×Buf., 1 μl KOD plus, 2 μl 25 mM MgSO4, 1.5 μl upstream and downstream primers, 1 μl template. The amplified fragment was connected to the T vector pMD19T (purchased from TaKaRa Company) to obtain pMD19T-pai.
[0026] The subcloning vectors pUC57-opai and pMD19T-pai were digested with endonucleases Pml I and Kpn I at 37°C for 1.5h, separated and recovered by 1% agarose gel electropho...
Embodiment 3
[0027] Embodiment 3 Preparation of recombinant Yarrowia lipolytica
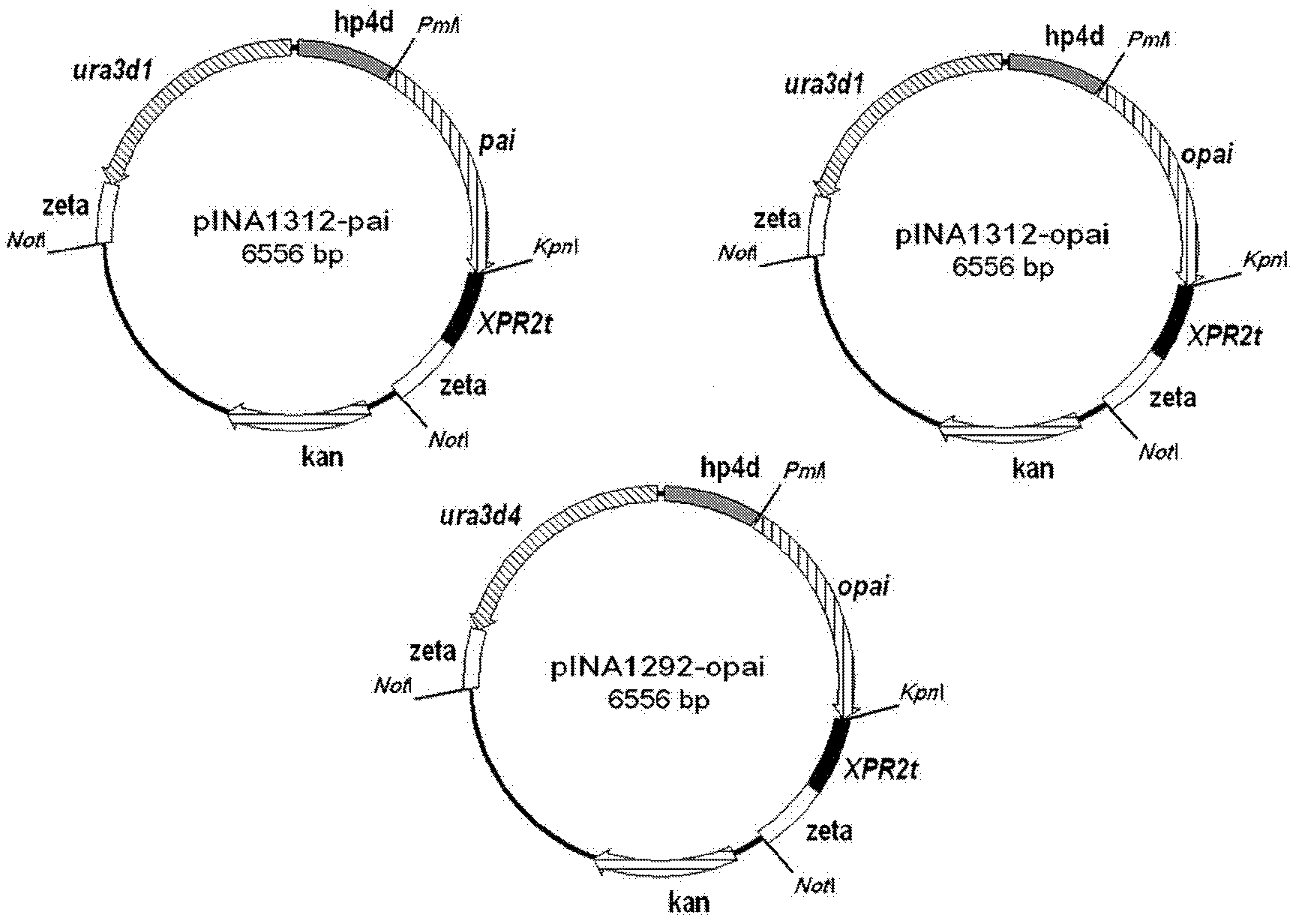
[0028] The three recombinant plasmids pINA1312-pai, pINA1312-opai and pINA1292-opai were digested with Not I at 37°C for 1.5h, separated and recovered by 1% agarose gel electrophoresis, and the expression unit fragment with the target gene was concentrated to 500ng / μl , Take 2μtg for transformation of Yarrowia lipolytica polh. The conversion process is as follows:
[0029] 1) Strain Y.lipolytica Polh was streaked on YPD solid medium, and cultured at 28°C for 12h;
[0030] 2) Pick a ring of Polh from the plate with an inoculation loop and resuspend in 1 mL TE solution;
[0031] 3) Centrifuge at 10,000 rpm to collect the bacteria, suspend the bacteria in 600 μl of 0.1 mol / l lithium acetate (pH6.0) buffer, bathe in 28°C water for 1 hour, and be careful not to shake the sample;
[0032] 4) After the water bath is over, centrifuge at 3000rpm for 2min, discard the supernatant;
[0033] 5) Gently resuspend the b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com