Application of tanshinone IIA in preparation of medicament for treating p53 mutational or deficient tumor
A tumor drug, tanshinone technology, applied in the field of application of tanshinone IIA in the preparation of tumor drugs for the treatment of p53 mutations or deletions, which can solve the problems of reduced tumor sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0012] Embodiment 1.MTT experiment
[0013] Experimental Materials:
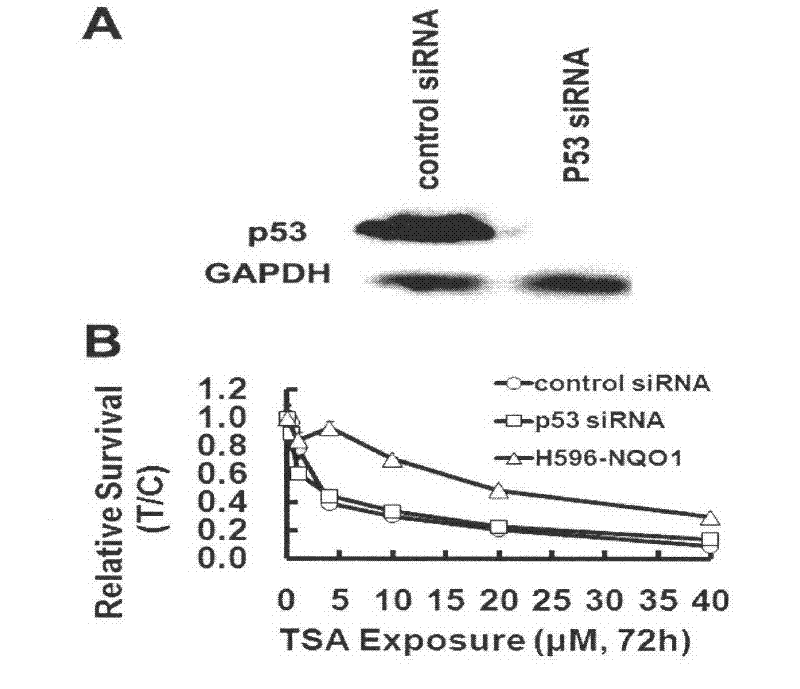
[0014] Human non-small cell lung cancer cells A549 cells and H596 cells were purchased from ATCC at 37°C, 5% CO 2 Under conventional conditions, the medium was RPMI-1640 (Gibco) containing 10% calf serum (PAA). The NQO1 gene was synthesized from the whole gene, and IRES2-EGFP was fused downstream to construct a lentiviral expression vector containing the NQO1-IRES-EGFP fragment. The 293T cells were used to package the lentivirus, the virus stock solution was concentrated, and the virus titer was determined. Wild-type H596 cells were infected to construct a H596 cell line (H596-NQO1) stably expressing NQO1. RNAiMAX transfection reagent, control siRNA, and P53 siRNA were purchased from Invitrogen. P53 antibody was purchased from Millipore.
[0015] experimental method:
[0016] 1. P53 gene silencing
[0017] A549 cells were seeded in a 96-well plate at a density of 8000 cells / well. After 18-24 hours, a...
Embodiment 2
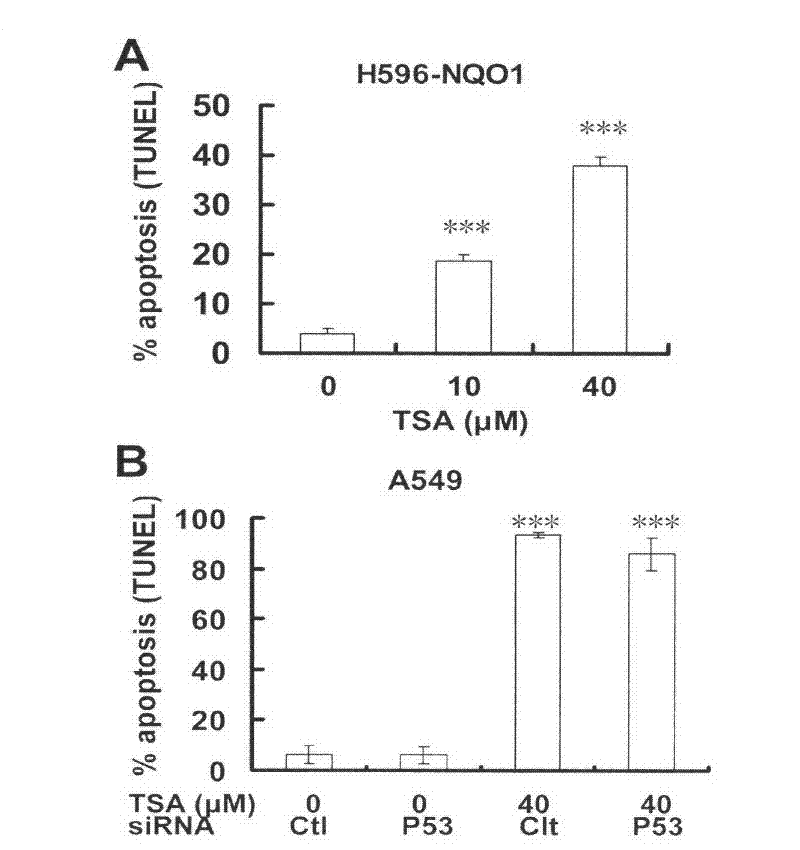
[0021] Embodiment 2.TUNEL experiment
[0022] Experimental material: with embodiment 1.
[0023] experimental method:
[0024] A549 cells or H596 cells were seeded in 6-well plates, and when the cell density reached 85% confluence, the A549 cells were administered. The experimental groups were as follows:
[0025] (1) H596-NQO1 cell grouping includes: TSA 0μM, 10μM, 40μM administration groups;
[0026] (2) A549 cell grouping: control siRNA transfection plus blank medium group
[0027] TSA 40μM administration group after control siRNA transfection
[0028] P53siRNA transfection plus blank medium group
[0029] TSA40μM administration group after P53siRNA transfection
[0030] The administration time was 48h. ApoBrdU DNA Fragmentation Assay Kit (BD) was used for TUNEL detection. The detection steps are as follows:
[0031] Fix cells → wash cells → TdT / BrdU labeled cells → wash cells → Anti-BrdU-FITC stained cells → wash cells → PI / Rnase treatment → flow cytometric analysi...
Embodiment 3
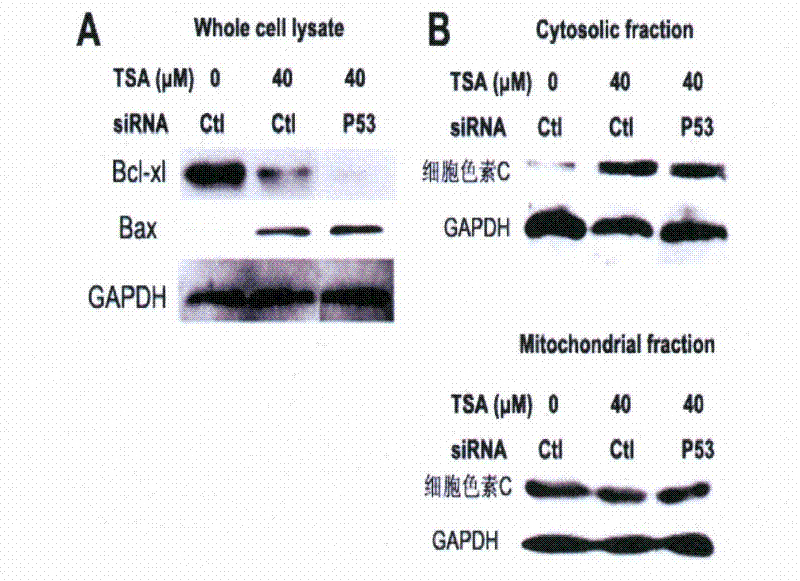
[0032] Example 3. Mitochondrial membrane potential detection
[0033] Experimental materials: A549 cells, JC-1 dye was purchased from KGI.
[0034] experimental method:
[0035] A549 cells at 1.5×10 4 Cells / well were seeded in 24-well plates, and the experimental groups were as follows
[0036] Control siRNA transfection plus blank medium group; control siRNA transfection TSA 40μM administration group;
[0037] Add blank medium group after P53siRNA transfection; TSA40μM administration group after P53siRNA transfection
[0038] After 24 hours of administration, remove the medium, wash the cells once with Hank's, add 3 μM JC-1 reagent and incubate at 37°C for 15 minutes, wash the cells twice with Hank's, and use synergy H1 multifunctional medium to determine the JC-1 polymer The red fluorescence emitted (excitation wavelength: 530nm, emission wavelength: 590nm) and the green fluorescence emitted by JC-1 monomer (excitation wavelength: 490nm, emission wavelength: 529nm) were...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com