Method for extracting metagenome DNA of compost and application thereof in identifying denitrifying bacteria
A technology of denitrifying bacteria and metagenomics, applied in the direction of recombinant DNA technology, DNA preparation, etc., can solve the problems of limiting the research and utilization of soil microbial resources and their genetic resources, time-consuming, poor accuracy, etc., to achieve short and efficient extraction , the effect of high application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1. Extraction of compost metagenomic DNA and its application in identifying denitrifying bacteria
[0053] 1. Production of chicken manure compost
[0054] 25kg of chicken manure, 12.3kg of sawdust and 13kg of water are mixed, the C / N of the mixture is 25-30, and the water content is 55-60%, and the mixture is fermented at room temperature (about 23° C.) for 20 days.
[0055] Chicken manure was obtained from the chicken farm of China Agricultural University, and sawdust was obtained from Yongfeng Woodware Factory, Haidian District, Beijing. The properties of the materials are shown in Table 1.
[0056] Table 1 Basic properties of compost raw materials
[0057]
[0058] Samples were taken on the 1st, 4th, 7th, 11th, 16th, and 20th days of fermentation for subsequent experiments.
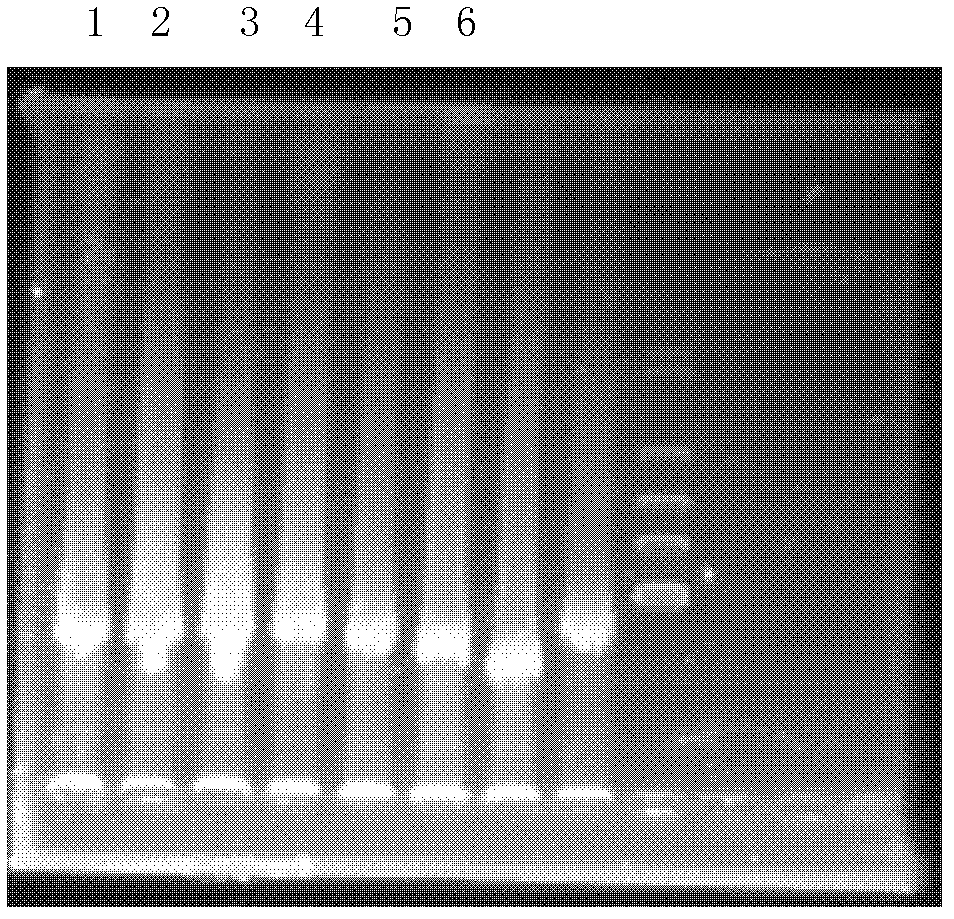
[0059] 2. Extraction of compost metagenomic DNA
[0060] 1. Weigh about 4g of chicken manure compost prepared in step 1 and place it in a 50ml centrifuge tube.
[0061] 2. Add 13...
Embodiment 2
[0084] Example 2. Extraction of compost metagenomic DNA and its application in identifying denitrifying bacteria
[0085] 1. Production of chicken manure compost
[0086] Same as step 1 in Example 1.
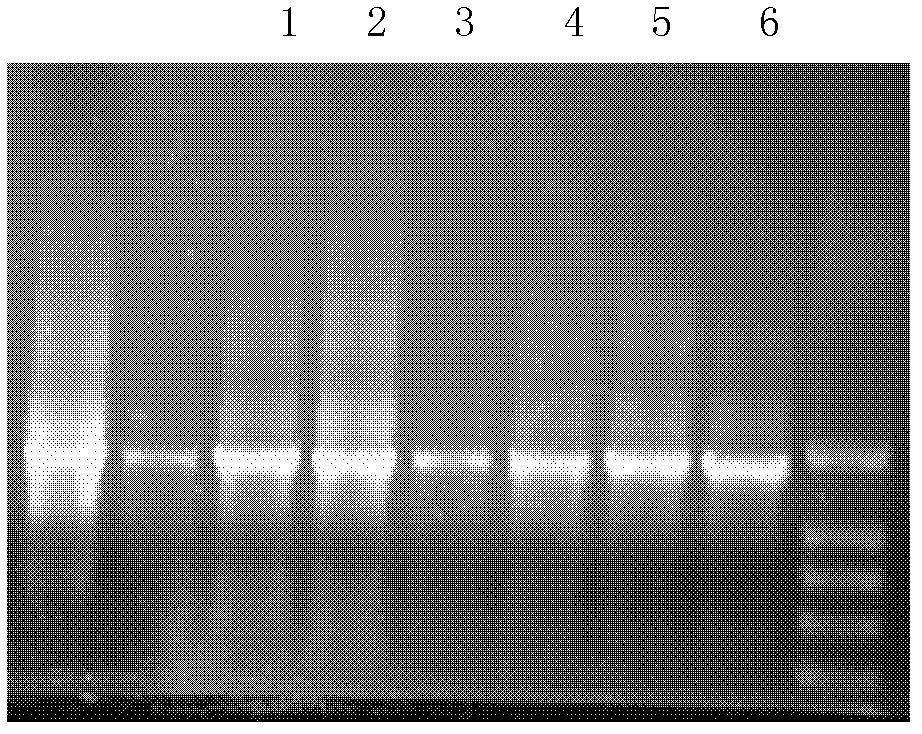
[0087] 2. Extraction of compost metagenomic DNA
[0088] 1. Weigh about 4g of chicken manure compost prepared in step 1 and place it in a 50ml centrifuge tube.
[0089] 2. Add 10ml of DNA extraction solution to the centrifuge tube in step 1, mix well and then perform physical lysis (repeat the following operation twice: liquid nitrogen treatment for 10 minutes, thawing in a 63°C water bath).
[0090] 3. After the liquid in the centrifuge tube in step 2 is slightly cooled, add 20 μl lysozyme (80 mg / ml), bathe in 36°C water for 25 minutes, and mix it upside down every once in a while (about 5 minutes).
[0091] 4. Add 100 μl of proteinase K (8 mg / ml) to the centrifuge tube in step 3, bathe in water at 36°C for 25 minutes, and mix by inverting up and down every once in a while (...
Embodiment 3
[0105] Example 3. Extraction of compost metagenomic DNA and its application in identifying denitrifying bacteria
[0106] 1. Production of pig manure compost
[0107] 25kg of pig manure and 3.1kg of sawdust are mixed, the C / N of the mixture is 25-30, and the water content is 55-60%, and the mixture is fermented at room temperature (about 23° C.) for 20 days.
[0108] Fresh pig manure was obtained from Shangzhuang pig farm in Beijing, and sawdust was obtained from Yongfeng Woodware Factory in Haidian District, Beijing. The properties of the materials are shown in Table 2.
[0109] Table 2 Basic properties of compost raw materials
[0110]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com