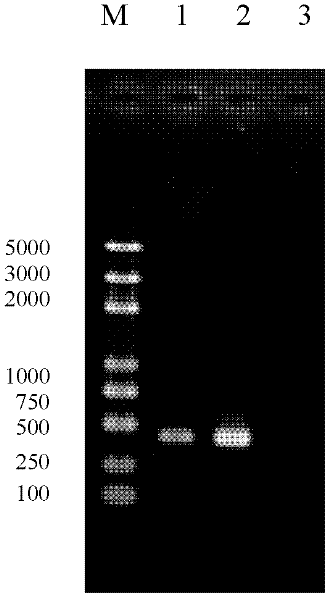
Method for assisting in detecting and identifying histamine-producing enterobacteria
A histamine-producing intestinal tract and auxiliary identification technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, recombinant DNA technology, etc., can solve pollution and other problems, and achieve timely quality control, easy identification, and simple methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Embodiment 1, design and preparation of primers
[0018] A specific primer pair (composed of primer LB1 and primer LB2) was designed according to the conserved region of the histidine decarboxylase coding gene (hdc gene) of various histamine-producing enterobacteria on NCBI. A universal primer pair (composed of primer B-for and primer B-for) for 16S rDNA of various histamine-producing bacteria was selected. Synthesize specific and universal primer pairs.
[0019] LB1 (sequence 1 of the sequence listing): 5'-AAGATACCCATTACTCCGTG-3';
[0020] LB2 (SEQ ID NO: 2 of the Sequence Listing): 5'-TCTACAGAGATCCGATCGAC-3'.
[0021] B-for (sequence 3 of the sequence listing): 5'-AGAGTTTGATCCTGGCTCAG-3';
[0022] B-rev (SEQ ID NO: 4 of the Sequence Listing): 5'-AAGGAGGTGATCCAGCCGCA-3'.
Embodiment 2
[0023] Example 2. Detection and identification of histamine-producing intestinal bacteria in fermented food
[0024] 1. Sample processing
[0025] Aseptically weigh 20 g of commercially available soy sauce koji, cut it into pieces under aseptic conditions, place it in a triangular flask filled with 180 ml of sterilized normal saline, mix well, and shake on a homogenizer for 10 min (the dilution is 10 -1 ); then carry out 10-fold serial dilution with sterilized normal saline, as each sample dilution (dilution is 10 successively -2 、10 -3 ,...).
[0026] 2. Isolation of intestinal bacteria from samples
[0027]Preparation of crystal violet neutral red bile salt glucose agar medium (VRBGA medium): 3.0g yeast extract, 7.0g peptone, 5.0g sodium chloride, 1.5g No. 3 bile salt, 10.0g lactose, 10.0g glucose, 0.03g neutral red, 0.002g crystal violet, and 15.0g agar were adjusted to 1000mL with distilled water, dissolved by heating, and adjusted to pH 7.4±0.2.
[0028] Spread the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com