Haemophilus parasuis real-time fluorescent quantitative PCR (polymerase chain reaction) detection method
A real-time fluorescence quantitative, Haemophilus suis technology, applied in the field of fluorescence quantitative PCR detection, can solve the problems of limited anti-interference ability of related bacteria and no effective verification of versatility.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0038] 1.4 Preparation of template DNA
[0039] Pick a single HPS colony and streak it on TSA agar medium containing 5% horse serum and 0.4 mg / mL NAD, place in 5% CO 2 Incubate at 37°C for 36 hours in the environment, elute the bacteria with normal saline, and extract bacterial DNA with a bacterial genome DNA extraction kit, and the extraction method is carried out according to the instructions.
[0040] 1.5 Qualitative PCR amplification
[0041] The designed HPS primers were used to amplify the extracted DNA template by PCR. The PCR reaction system was 50 μL, including 12 μL of PCR Taq Mix, 1 μL of upstream and downstream primers (25 pmol / μL), 5 μL of template DNA, and 31 μL of ultrapure water. The reaction conditions were pre-denaturation at 95°C for 5min; 35 cycles of 95°C for 20S, 60°C for 20S, 72°C for 20S, and finally 72°C for 5min.
[0042] 1.6 Preparation of standard positive plasmid
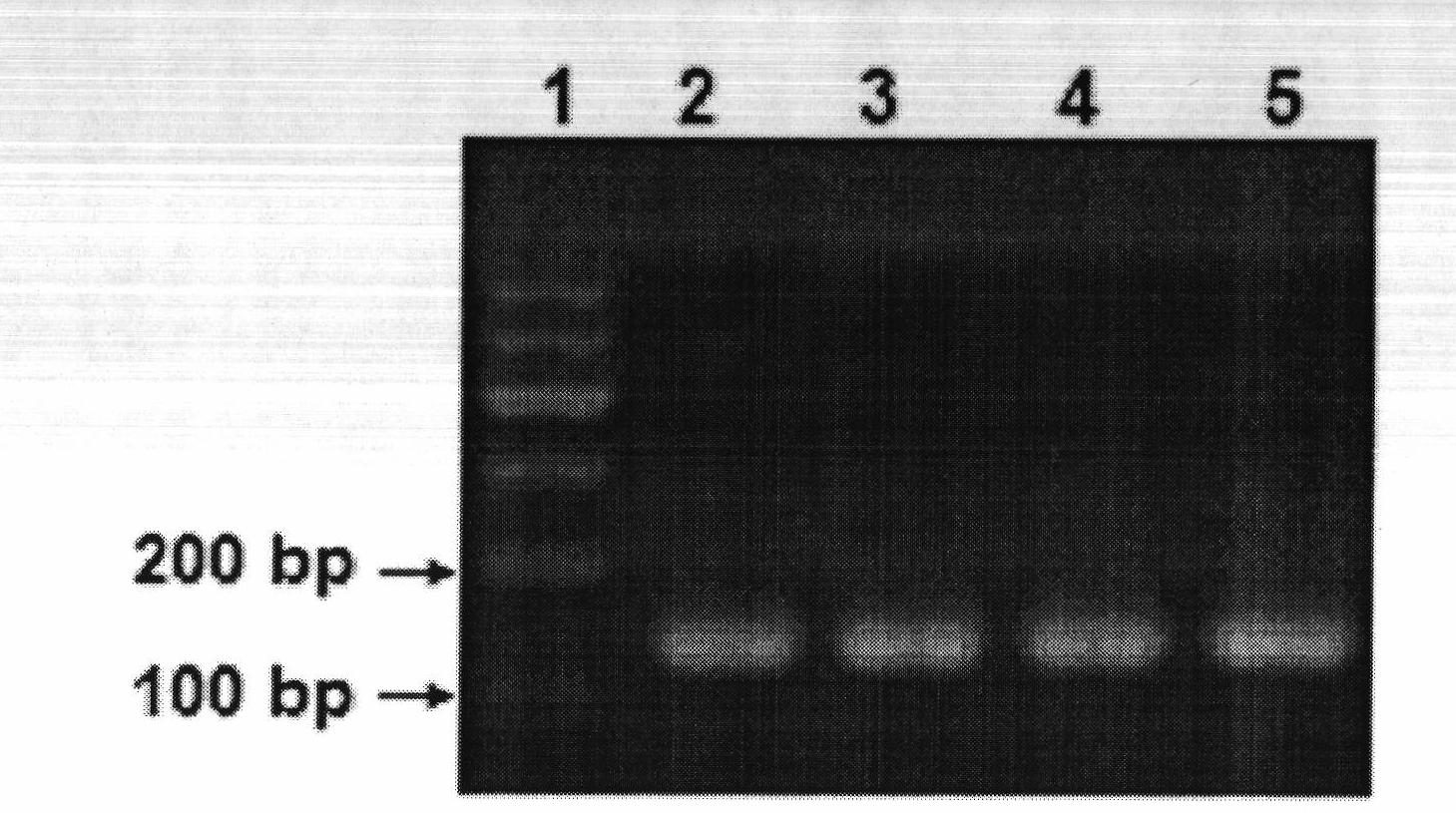
[0043] After the PCR product was electrophoresed on 2% agarose gel, the PCR produ...
Embodiment
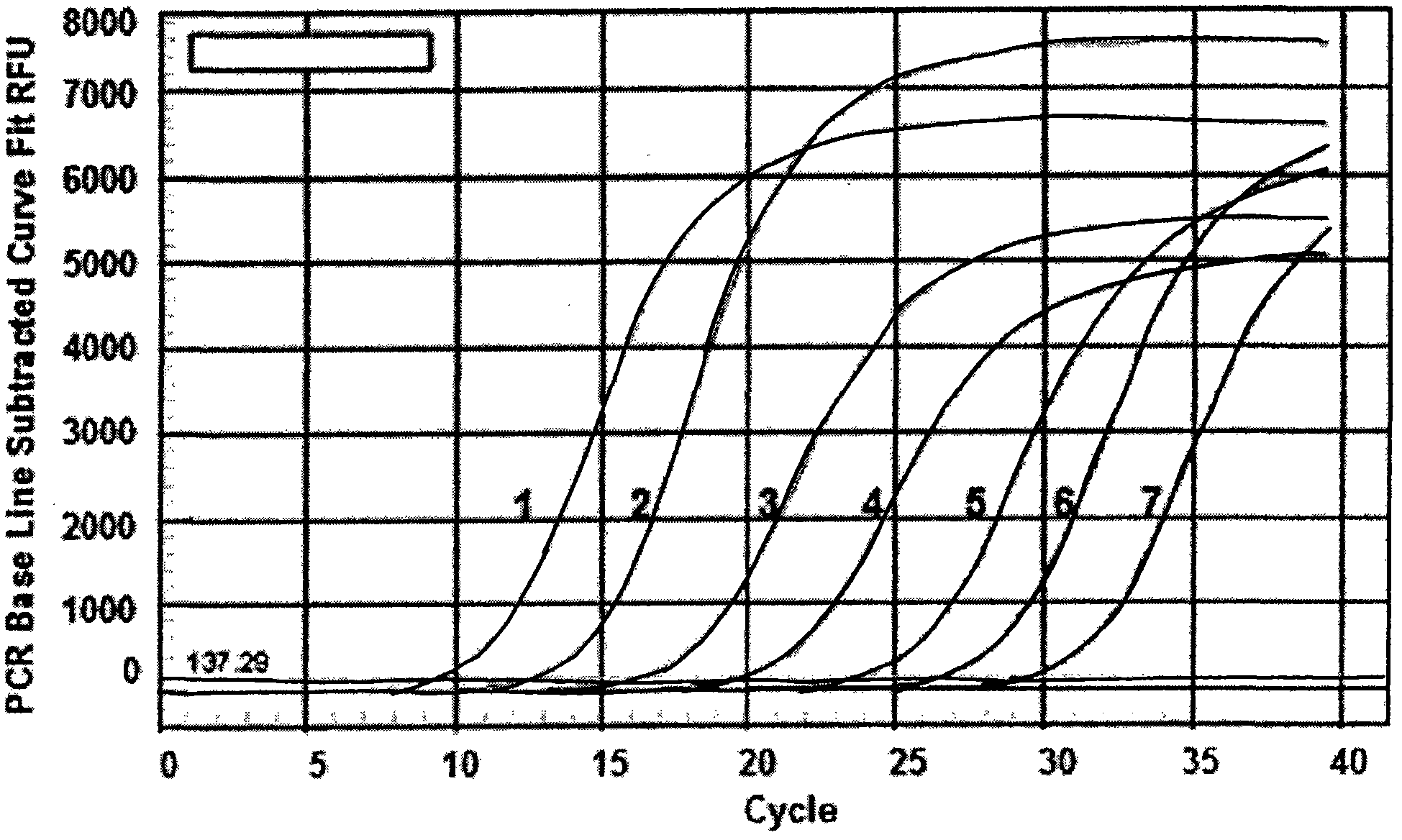
[0065] Example Detection of clinical samples
[0066] The established real-time fluorescent quantitative PCR detection method for Haemophilus parasuis (method A) was used to detect 10 clinically collected pericardial fluids and joint fluids suspected of HPS infection, and they were separated and cultured from bacteria (method B) and conventional PCR (method B) C) for comparison.
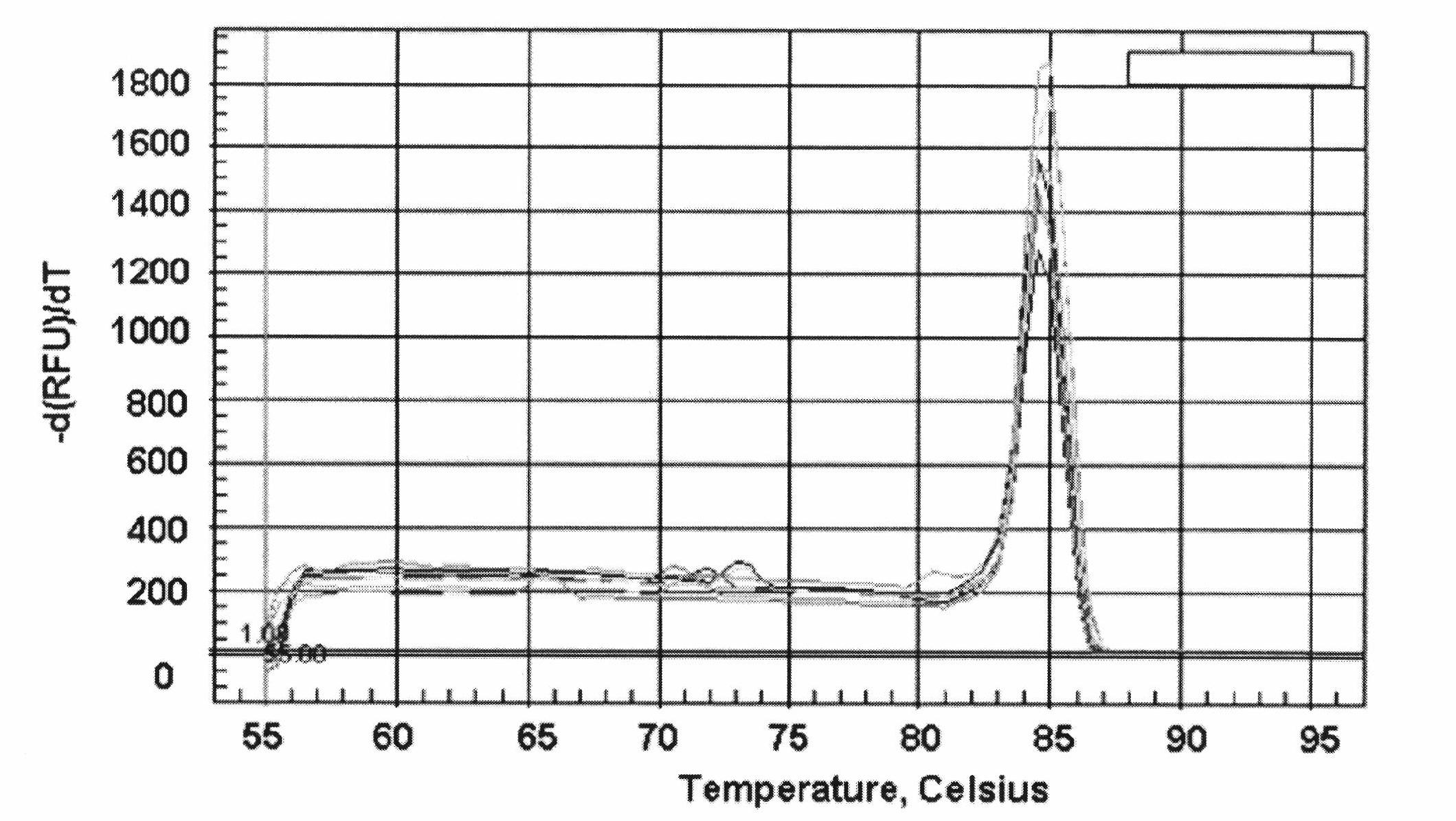
[0067] Such as Figure 7 As shown, the specific steps of the real-time fluorescent quantitative PCR detection method for Haemophilus parasuis are as follows:
[0068] DNA sample preparation: take the sample to be tested for extraction and purification of total DNA;
[0069] Pericardial fluid treatment: draw 10 microliters of pericardial fluid into an EP tube containing 100 microliters of ultrapure water, boil for 10 minutes, centrifuge at 12,000 rpm for 5 minutes, and take 3 microliters of the supernatant as a DNA template.
[0070] Joint fluid treatment: pipette 10 microliters of joint fluid int...
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com