Method for obtaining biomass conversion related genes from anaerobic fermentation system
A technology for anaerobic microorganisms and biomass, applied in the fields of biotechnology and microorganisms, which can solve the problems of lack of phylogenetic evolution or taxonomic status, inability to obtain target microorganisms, and inability to obtain pure cultures.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0066] As an embodiment of the present invention, the method of the present invention includes: (1) starting from the anaerobic fermentation system, especially starting from the stalk anaerobic fermentation system, extracting and purifying the metagenomic DNA of 30-48kb, after Ligation, packaging and transfection to establish the Fosmid metagenomic library; (2) using the hydrolysis circle color method to screen the Fosmid library for enzymes related to biomass conversion such as endoglucanase, exoglucanase, β- Positive clones of glucosidase and endoxylanase; (3) After the positive clones screened were mixed in equal proportions, the Fosmid DNA was extracted and sequenced by 454 sequencing, and the genes encoding the corresponding enzymes were compared with the existing database Compare to identify whether it is a new gene with less than 85% similarity to the protein sequence encoded by the reported gene, and at the same time obtain the (upstream and downstream) regulatory seque...
Embodiment 1
[0072] Example 1, Construction of Fosmid Metagenomic Library in Biogas Fermentation System
[0073] 1. Genomic DNA extraction
[0074] Take 8mL of biogas slurry (taken from a biogas digester with straw and pig manure as raw materials for anaerobic fermentation at 40°C), centrifuge at 8000g for 5min, take the precipitate and wash it with 20mL of PBS buffer (137mM NaCl, 2.7mMKCl, 1.5mM KH 2 PO 4 , 8.1 mM Na 2 HPO 4 , pH 7.4) and washed 3 times. Resuspend the pellet with 7.68mL DNA extraction buffer (100mM Tris-HCl (pH 8.0), 100mM EDTA, 100mM Na 3 PO 4 , 1.5M NaCl, 1% (w / v) CTAB). Add proteinase K (final concentration 1 mg / mL) and SDS (final concentration 1% (w / v)) and incubate at 55° C. for 20 minutes, and then incubate at 70° C. for 10 minutes. After the crude lysate was centrifuged at 17,000 g for 10 min, the supernatant was collected. The supernatant was extracted twice with phenol:chloroform:isoamyl alcohol (25:24:1) and then once with chloroform:isoamyl alcohol (24:...
Embodiment 2
[0084] Example 2, Screening of Positive Clones Containing Genes of Enzymes Related to Biomass Conversion
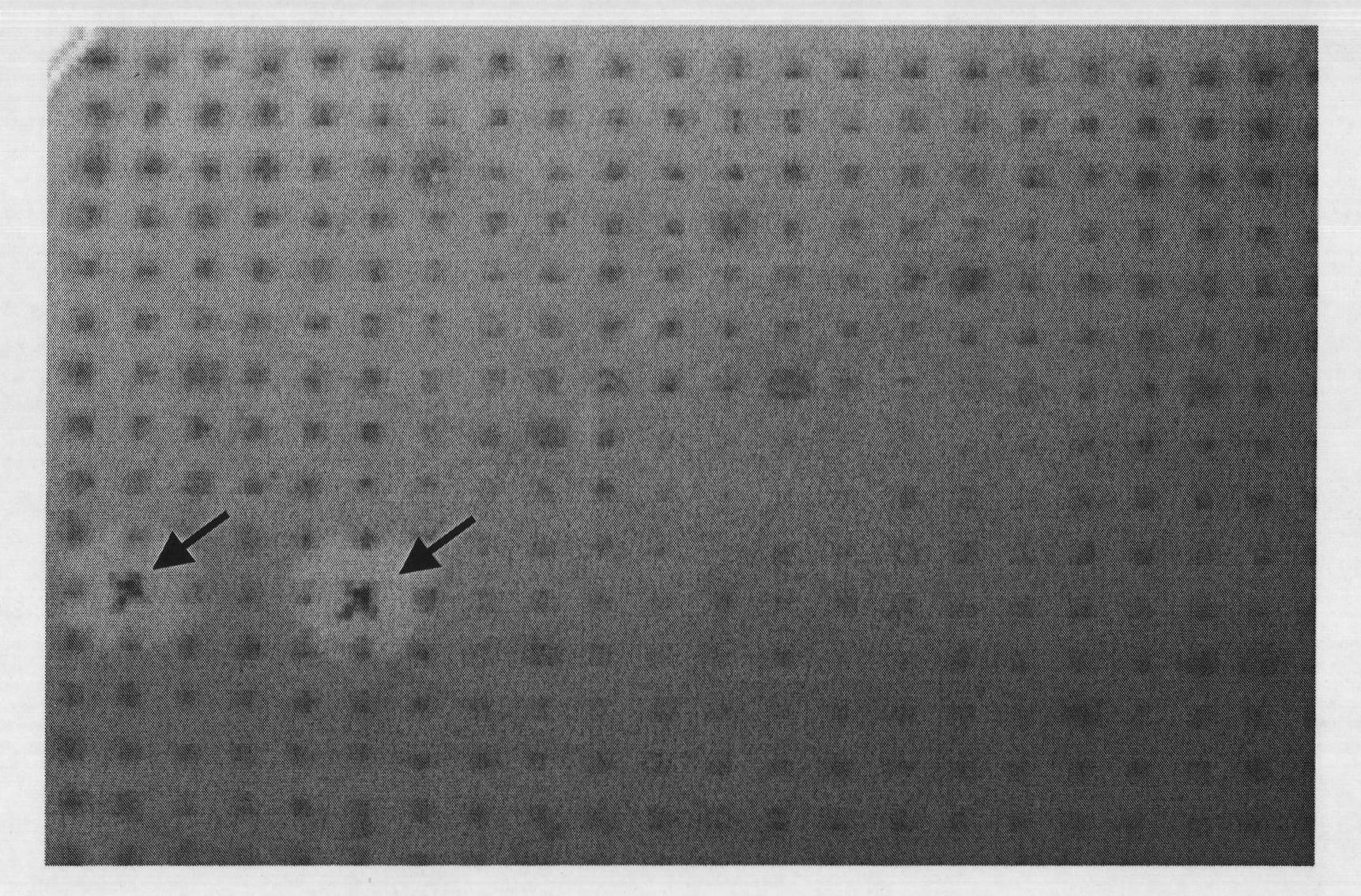
[0085] Fosmid clones were replicated from 384-well plates to LB plates containing 0.5% (w / v) carboxymethylcellulose sodium (CMC-Na) (containing 12.5 μg / mL chloramphenicol) using 384-well plate inoculation pins. After incubation at 37°C for 20 hours, the cells were stained with Congo red. After pouring 0.2% (w / v) Congo red staining solution and staining for 30 minutes, wash with sodium chloride solution (1mol / L) at least twice. The endoglucanase-positive clones have obvious yellow hydrolysis circles on the side of the colonies. From the 281 preserved 384-well plates, select one of them (No. BF009) to screen endoglucanase according to the above method, and screen 2 endoglucanase positive clones with yellow hydrolysis circles, as figure 1 indicated by the middle arrow. Using this method, 341 positive clones were screened from the Fosmid library containing 100,000 clones. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com