Molecular marker linked with gummy stem blight resistance gene Gsb-2 and application thereof
A molecular marker, gsb-2 technology, applied in the field of agricultural biology, can solve the problems of individual plants that are difficult to screen for disease resistance genes, unsuitable disease conditions, affecting selection efficiency, etc., so as to improve the selection accuracy, shorten the breeding cycle, select targeted effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
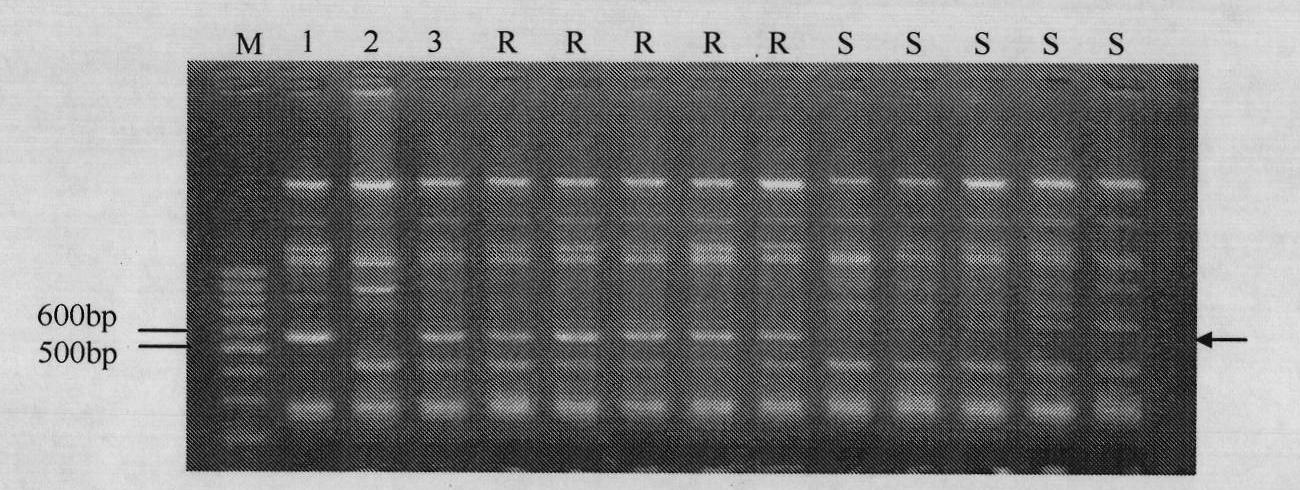
[0045] Example 1: ISSR molecular marker ISSR-57 linked to muskmelon blight resistance gene Gsb-2 560 the acquisition
[0046] 1) F 1 , F 1 F 2 . By planting parents, F 1 Generation and F 2 generation groups. It is artificially inoculated and the incidence of each individual plant is counted, F 2 The population statistics of generation segregation showed that in the population of 134 strains, 83 strains were disease-resistant and 36 strains were susceptible, which conformed to the inheritance pattern of 3:1 single dominant gene.
[0047] 2) Inoculation of creeping blight and disease classification: the method of spraying inoculation with conidia suspension and 5-level disease classification (Zhang et al., 1997), spraying the spore suspension with a micro-sprayer, the concentration of the spore suspension is 5 × 105 pcs mL -1 , Spray until the leaves of the plants start to drip. After inoculation, use a small shed to keep moisture, and the relative humidity is above 90...
Embodiment 2
[0056] Example 2: Amplification detection of the parents by primer ISSR-57
[0057] Using the parental PI 157082 and the genomic DNA of Baipicrisp as templates, ISSR-PCR amplification was carried out with primer ISSR-57, and the ISSR-PCR reaction system was: 10×buffer (without Mg 2+ ) 2.0μL; dNTP (2mmol L -1 ) 2.0 μL; MgCl 2 (25mmol·L -1 ) 1.5μL; Primer (10mmol L -1 ) 1.0 μL; template DNA (30ng·μL -1 ) 1.0 μL; Taq polymerase (5U·μL -1 ) 0.2 μL; ddH 2 O 12.3 μL, a total of 20 μL. The PCR amplification reaction program is: pre-denaturation at 94°C for 5 minutes, then denaturation at 94°C for 30 seconds, annealing at 55°C for 45 seconds, extension at 72°C for 2 minutes, and 35 cycles of extension at 72°C for 10 minutes, and storage at 4°C. The entire reaction procedure is about 2.5h.
[0058] The amplified product was mixed with 1.5 μL bromophenol blue (0.25%), and the sample was spotted on a medium containing 0.5 tg·mL -1 In 1.5% agarose gel of EB, with 1×TAE as the ele...
Embodiment 3
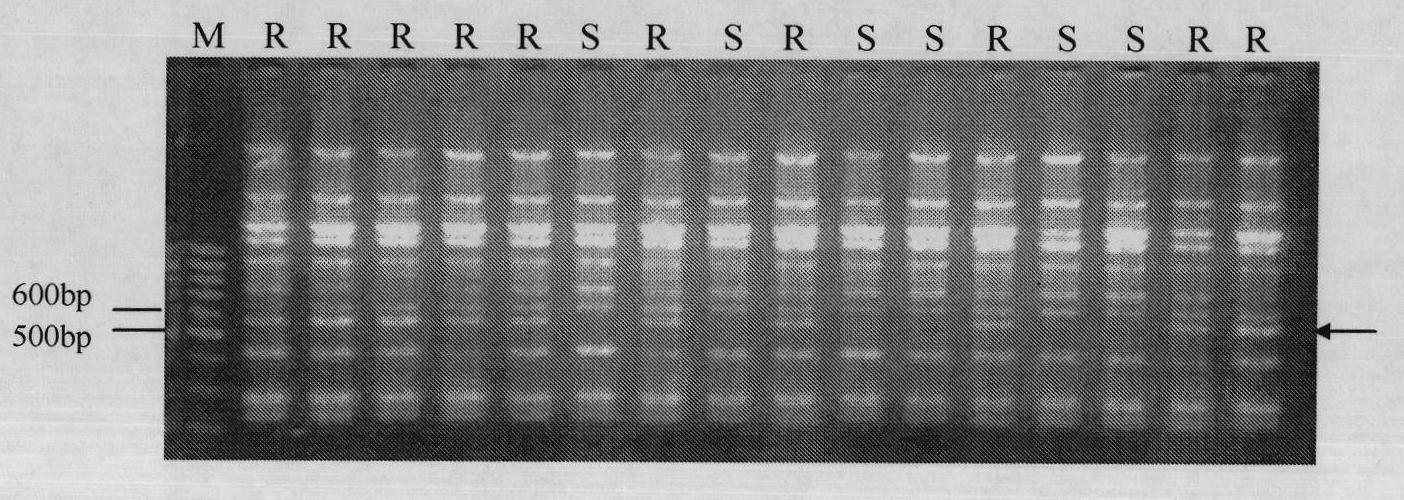
[0059] Example 3: Primer ISSR-57 to F 1 amplification detection
[0060] Take Hybrid F 1 Genomic DNA of Genomic DNA was used as a template, ISSR-PCR amplification and electrophoresis detection were carried out with primer ISSR-57, the amplification and detection method was: ISSR-PCR reaction system was: 10×buffer (excluding Mg 2+ ) 2.0μL; dNTP (2mmol L -1 ) 2.0 μL; MgCl 2 (25mmol·L -1 ) 1.5μL; Primer (10mmol L -1 ) 1.0 μL; template DNA (30ng·μL -1 ) 1.0 μL; Taq polymerase (5U·μL -1 ) 0.2 μL; ddH 2 O 12.3 μL, a total of 20 μL. The PCR amplification reaction program was as follows: pre-denaturation at 94°C for 5 min, followed by denaturation at 94°C for 30 s, annealing at 55°C for 45 s, extension at 72°C for 2 min, and 35 cycles, further extension at 72°C for 10 min, and storage at 4°C. The amplified product was mixed with 1.5 μL bromophenol blue (0.25%), and the sample was spotted on a medium containing 0.5 μg·mL -1 In 1.5% agarose gel of EB, with 1×TAE as the electro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com