Epoxide hydrolase and preparation method thereof
A technology of epoxide and hydrolase, which is applied in the field of genetic engineering, can solve problems such as temperature sensitivity, loss of activity, and enzyme instability, and achieve the effects of improving temperature stability, increasing service life, and high enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
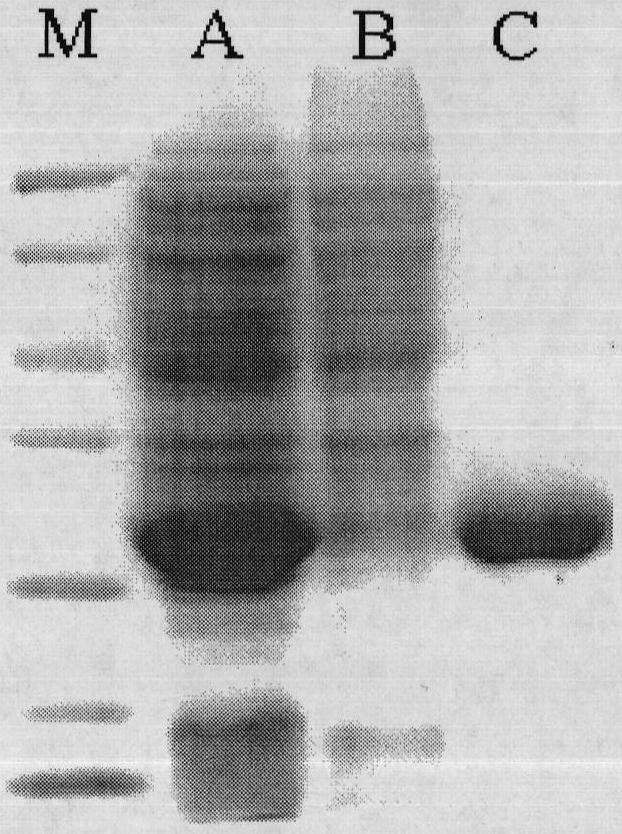
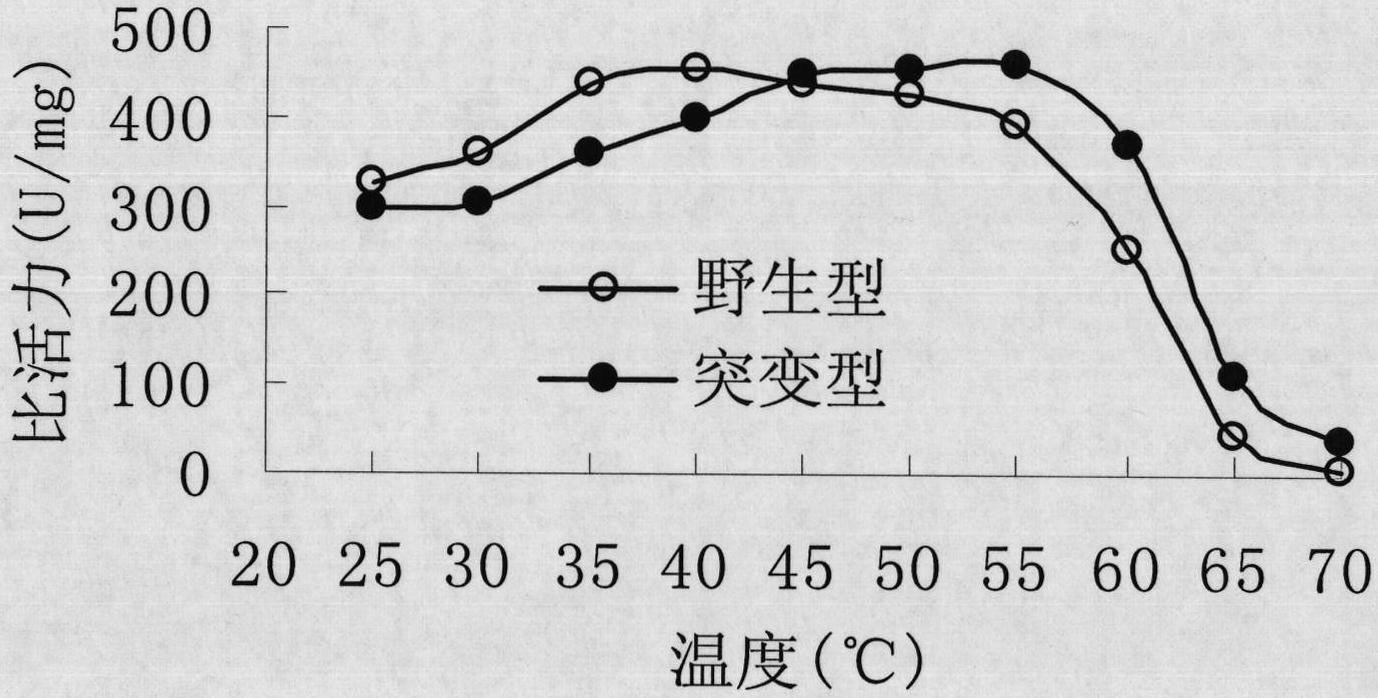
Image
Examples
Embodiment 1
[0041] Embodiment 1: Obtaining of Rhodococcus CGMCC1756 epoxide hydrolase gene
[0042] Rhodococcus CGMCC1756 was inoculated into the seed medium (1% glucose, 0.5% peptone, 0.5% beef extract, 1% sodium chloride, pH7.5), and cultured with shaking at 30°C for 24h. Genomic DNA of Rhodococcus CGMCC1756 was extracted with EZ-10 Column Genomic DNA Extraction Kit (purchased from Bio Basic Inc.) according to the instructions. According to the nucleotide sequence (accession number DQ471957) of Rhodococcus opacus ML-0004 (Rhodococcus opacusML-0004) epoxide hydrolase disclosed in the U.S. GenBank database, two primers (i.e. primer 1 and primer 2) were designed, primer 1 and The nucleotide sequences of primer 2 are respectively shown in SEQ ID NO: 1 and SEQ ID NO: 2. Using the extracted Rhodococcus CGMCC1756 genomic DNA as a template, using primers 1 and 2, a DNA fragment of about 800 bp was obtained by a commonly used PCR amplification technique. The PCR reaction system is: 40 μl of do...
Embodiment 2
[0043] Example 2: Construction of an expression system for epoxide hydrolase from Rhodococcus CGMCC1756
[0044] According to the sequencing result of Rhodococcus CGMCC1756 epoxide hydrolase gene in embodiment 1, redesign two primers (being primer 3 and primer 4), the nucleotide sequence of primer 3 and primer 4 is respectively as SEQID NO:5 and Shown in SEQ ID NO:6. Primer 3 contains Nco I restriction endonuclease site, 6 polyhistidine sequences and initiation codon. Primer 4 contains a Bam HI restriction endonuclease site and a stop codon. Using the Rhodococcus CGMCC1756 genomic DNA extracted in Example 1 as a template, using primers 3 and 4, a nucleotide fragment of about 800 bp was obtained by PCR amplification. The PCR reaction system is: 40 μl of double distilled water, 5 μl of 10×PCR buffer, 1 μl of 10 mmol / L dNTPs, 1 μl of 10 mmol / L primer 3, 1 μl of 10 mmol / L primer 4, 1 μl of Rhodococcus CGMCC1756 genomic DNA, 1 μl of Taq enzyme, A total of 50 μl of the reaction s...
Embodiment 3
[0046] Embodiment 3: Site-directed mutation of Rhodococcus CGMCC1756 epoxide hydrolase gene
[0047] The 163rd tyrosine (Tyr, corresponding codon is TAC) of the main amino acid sequence of Rhodococcus CGMCC1756 epoxide hydrolase is site-directed mutation to phenylalanine (Phe, corresponding codon is TTC or TTT). TTC and TTT are synonymous codons, and they all encode phenylalanine. The present invention uses the TTC codon as an example to design mutation primers (i.e., primer 5 and primer 6). The nucleotide sequences of primer 5 and primer 6 are as follows: Shown in SEQ ID NO: 7 and SEQ ID NO: 8. PCR amplification was performed using the recombinant vector pTrc99A-EH in Example 2 as a template, and primers 5 and 6 as mutation primers. The PCR reaction system is: 40 μl of double distilled water, 5 μl of 10×PCR buffer, 1 μl of 10 mmol / L dNTPs, 1 μl of 10 mmol / L primer 5, 1 μl of 10 mmol / L primer 6, 1 μl of recombinant vector pTrc99A-EH, 1 μl of Taq enzyme, A total of 50 μl of t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com