PIK3CA gene mutation detection probe, detection liquid phase chip and detection method thereof
A liquid phase chip and detection method technology, applied in the field of molecular biology, can solve the problems of poor specificity and high false positive rate of real-time fluorescent quantitative PCR detection technology, achieve uniform reaction conditions, avoid high false positive rate, and improve sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Preparation of liquid-phase chip for detection of PIK3CA gene mutation
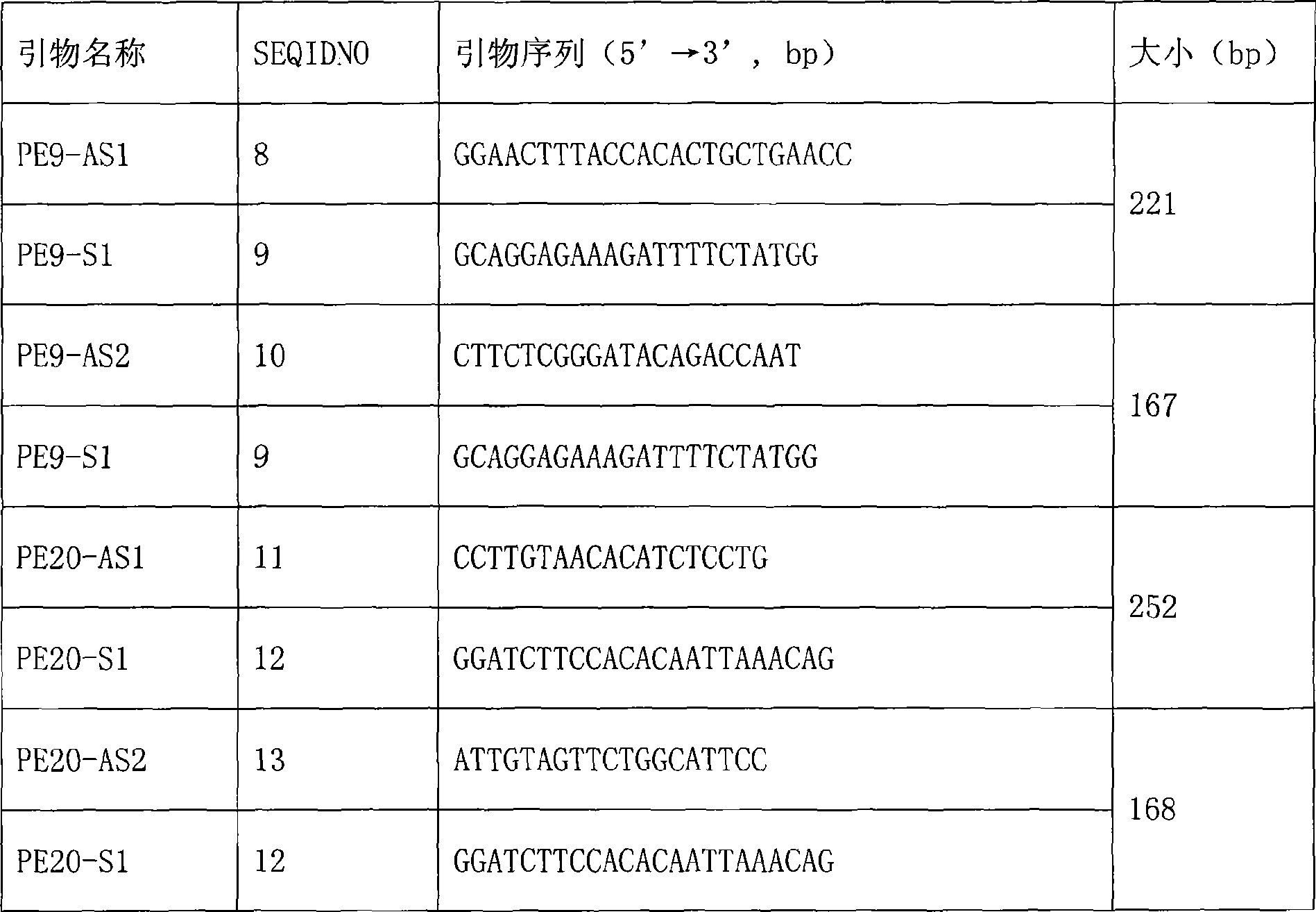
[0057] 1. Probe sequence design and microsphere coating
[0058] Specific oligonucleotide probes were designed for the wild-type and mutant sequences of exons 9 and 20 of the PIK3CA gene. The 5' end of the probe is an amino group followed by a 10 T spacer. The probe was synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd. Probes were coupled with different color-coded microspheres (purchased from Luminex) through covalent bonding (coating process).
[0059] The specific steps of each microsphere coating are as follows:
[0060] (1) Take the microsphere mother solution (purchased from Luminex Company) and vibrate on a vortex instrument to form a microsphere suspension;
[0061] (2) Take out 8ul microsphere mother solution, containing 0.8×10 5 —1.2×10 5 microspheres into a 0.5ml centrifuge tube;
[0062] (3) Centrifuge at 10,000rpm for 2min, discard the supernatant;
[0...
Embodiment 2
[0081] Using the PIK3CA gene mutation detection liquid chip in Example 1 to detect breast cancer tissue samples
[0082] 1. Preparation of samples to be tested
[0083] Extraction of DNA from breast cancer tissue samples: take 5-50 mg of tissue samples after breast cancer surgery, grind them, and wash them twice with PBS solution of pH 7.4; the washed tissue samples are resuspended in 1ml of digestive solution (50mmol / L Tris, 1mmo / L Na 2 E DTA, 0.5% Tween-20, 200ug / ml proteinase K 200, pH8.5), digested in a water bath at 55°C for 1 hour, inactivated proteinase K in a water bath at 99°C for 15 minutes; centrifuged at 12000 rpm for 10 minutes; took the supernatant, DNA samples for PCR reaction were obtained by extraction with phenol-chloroform-isoamyl alcohol and ethanol precipitation. DNA can also be extracted by micro spin column method;
[0084] 2. PCR amplification and enzyme digestion enrichment of the samples to be tested:
[0085] (1) PCR amplification and enzyme dig...
Embodiment 3
[0171] Example 3, PIK3CA gene mutation detection liquid chip detection sensitivity experiment
[0172] In order to test the detection sensitivity of this liquid-phase chip, the ability to detect mutant DNA in PIK3CA wild-type DNA was designed, that is, to investigate the minimum ratio of mutant DNA that this liquid-phase chip system can detect. The copy numbers of wild-type DNA and mutant DNA in each group of samples in this experiment are shown in Table 2. Each set of experiments was repeated. The steps of the first round of PCR, enzyme digestion reaction, second round of PCR reaction and luminex detection of the samples in this example are the same as those described in Example 2.
[0173] Table 3 Sensitivity test results of liquid-phase chip for PIK3CA gene mutation detection
[0174] wild-type DNA
copy number mutant DNA
copy number mutant DNA
The proportion E542K-P
(MFI) E545K-P
(MFI) E545D-P
(MFI) H1047R-P
(MFI) 500 5 1% ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com