Method for detecting protein content difference
A technology of protein content and determination method, which is applied in the field of relative protein content, can solve problems such as inconvenient and effective detection methods and cumbersome determination, and achieve the effect of less reagent consumption and low sample consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
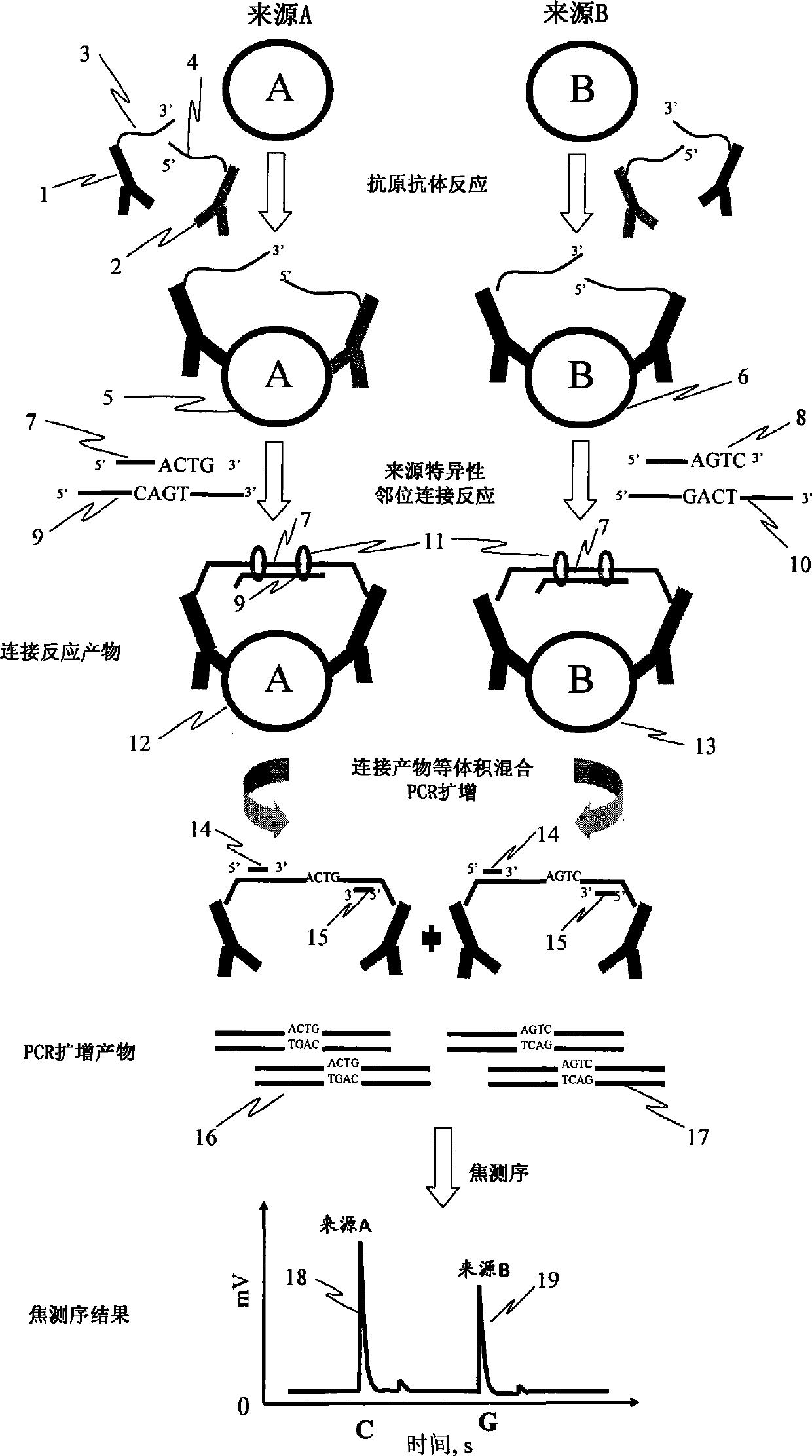
[0039] Embodiment 1: the determination of the relative content of interleukin 2 (IL-2) of two kinds of different sources
[0040] 1. Experimental materials: recombinant human interleukin-2 (Peprotech, USA); biotinylated anti-human interleukin-2 polyclonal antibody (R&D System, USA); maleimide-modified streptavidin (Sigma, USA); Magnetic beads (DynabeadsM-280-strptavidin, Dynal AS, Oslo, Norway); oligonucleotide single-stranded (Invitrogen synthesis, Shanghai) Probe 1: 5'-SH-ttttt ttatg tggtc tatgt cgtcg ttcgc tagta gttcc tgggc tgcac- 3'; Probe 2: 5'-P-tcgaggcgta gaatt ccccc gatgc gcgct gttct tactc atttt t-SH-3'; Sequence tag: 5'-P-agt tgc cat ctg tcgatc-3', 5'-P-agt tgc cat ctg tca ctg-3'; linking single strand: 5'-acgcc tcgac agtga cagat ggcaa ctgtgcagcc cttt-3', 5'-acgcc tcgag atcga cagat ggcaa ctgtg cagcc cttt-3'; amplification primer: 5'-Bio -atgtggtcta tgtcg tcgtt cg-3', 5'-tgagt aagaa cagcg cgcat-3'; annealing primer: 5'-ggggg aattc tacgcctcga-3'.
[0041] 2. Experimen...
Embodiment 2
[0052] Example 2: Determination of protein content differences by directly labeling source-specific sequences on DNA molecules linked to antibody proteins
[0053] In this example, the figure 1 The source-specific base sequences in DNA single strands 7 and 8 are marked in DNA sequence 3 or 4 respectively, that is, the DNA sequences in antigen-antibody-DNA complexes 5 and 6 formed after the antigen-antibody reaction are different and contain source-specific sexual sequence. When performing the source-specific proximity ligation reaction, it is only necessary to add a single-stranded DNA 9 complementary to the sequences in 3 and 4 for the ligation reaction. The rest of the steps are the same as figure 1 unanimous.
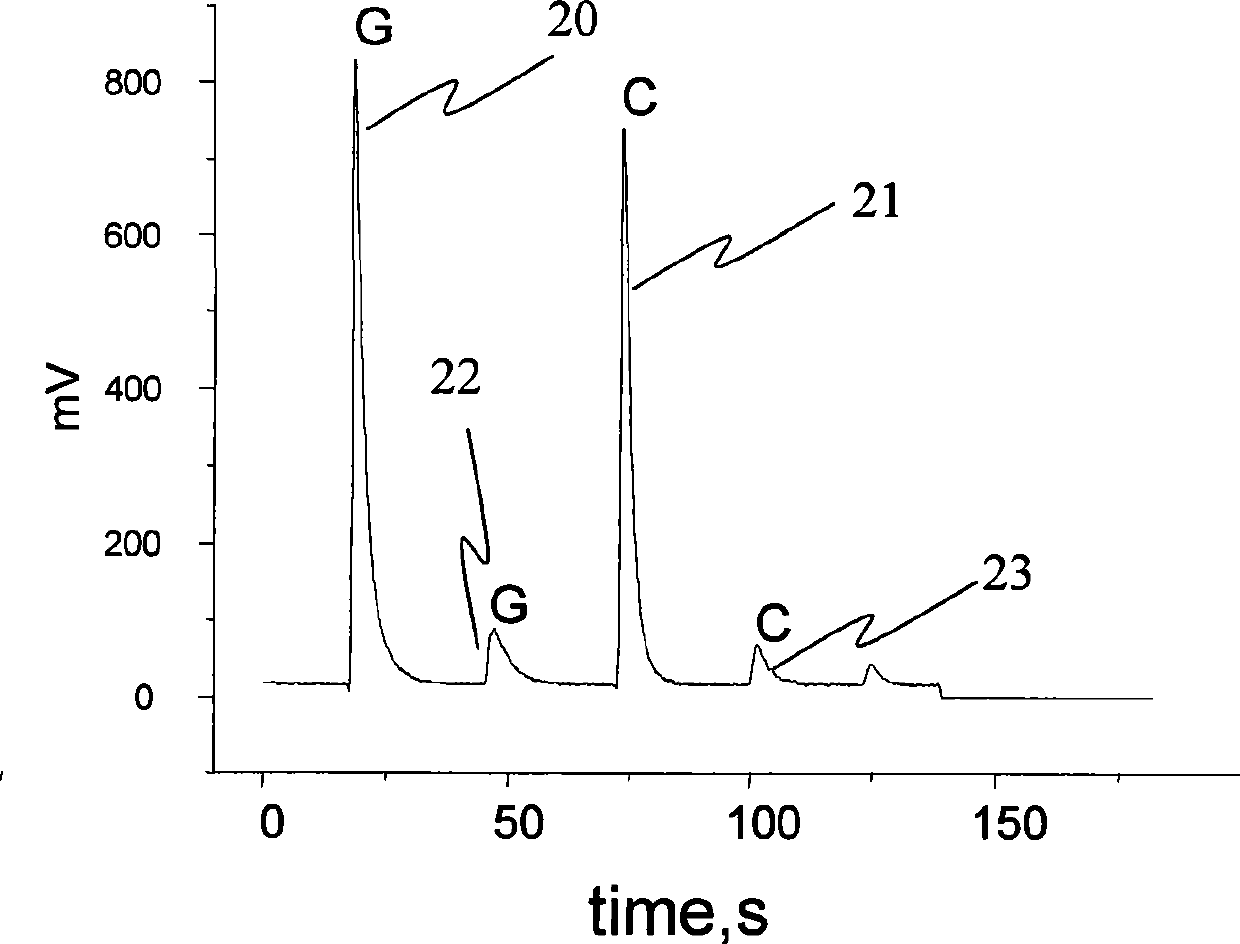
[0054] In this example, DNA with different sequences needs to be labeled with the same antibody to distinguish proteins from different sources. The determination procedure is the same as that in Example 1. Since the content of IL2 from the two sources is equal, t...
Embodiment 3
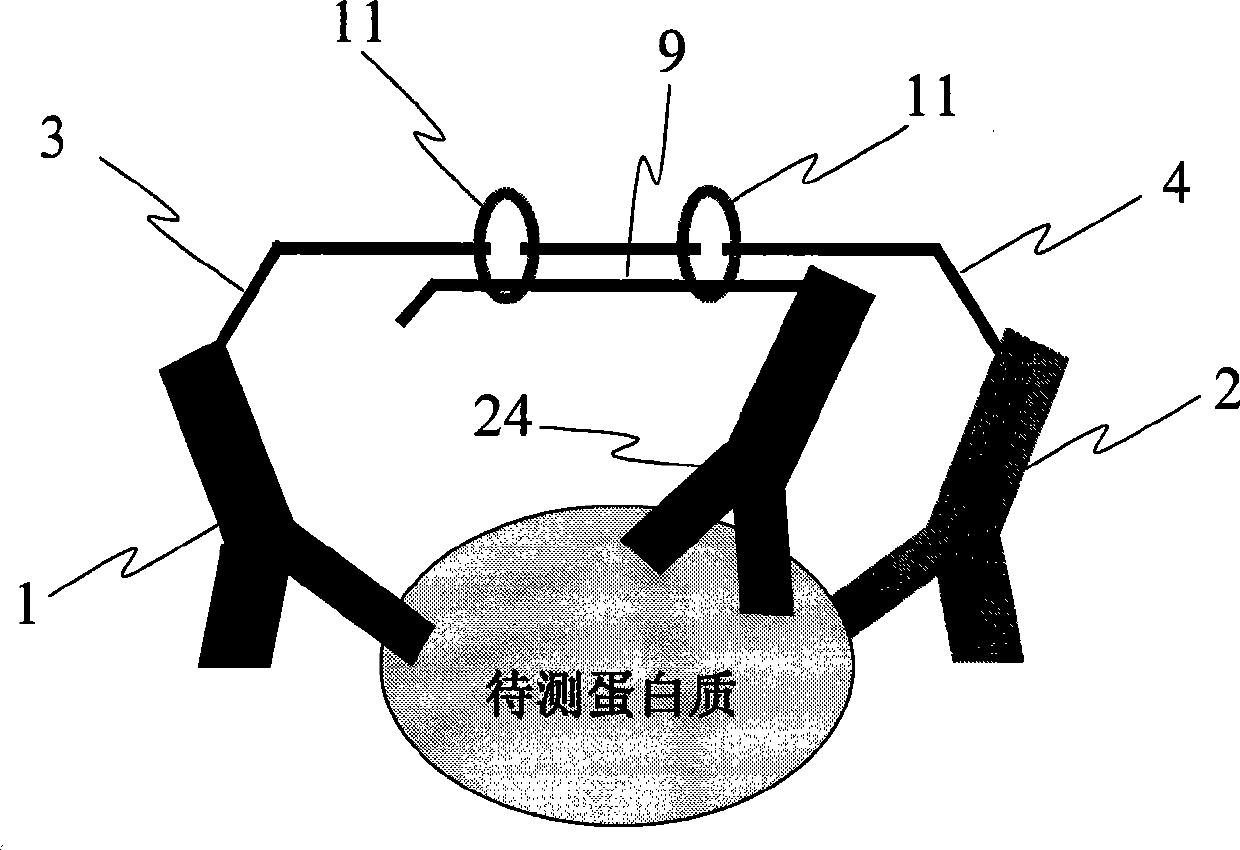
[0055] Example 3: The method of adding a third antibody to reduce the background signal of the reaction and improve the sensitivity of the assay
[0056] In this embodiment, at least three antibody-DNA composite probes are prepared, and the antibodies are antibodies to the same protein. Such as image 3 shown, except figure 1 In addition to the antibodies 1 and 2, an antibody 24 should be added, and the adjacent linked DNA single strand (9) containing the source-specific sequence should be connected with the antibody 24. After the antigen-antibody reaction occurs, the ligation reaction occurs under the action of ligase 11. This results in increased specificity and reduced background in the ligation reaction.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com