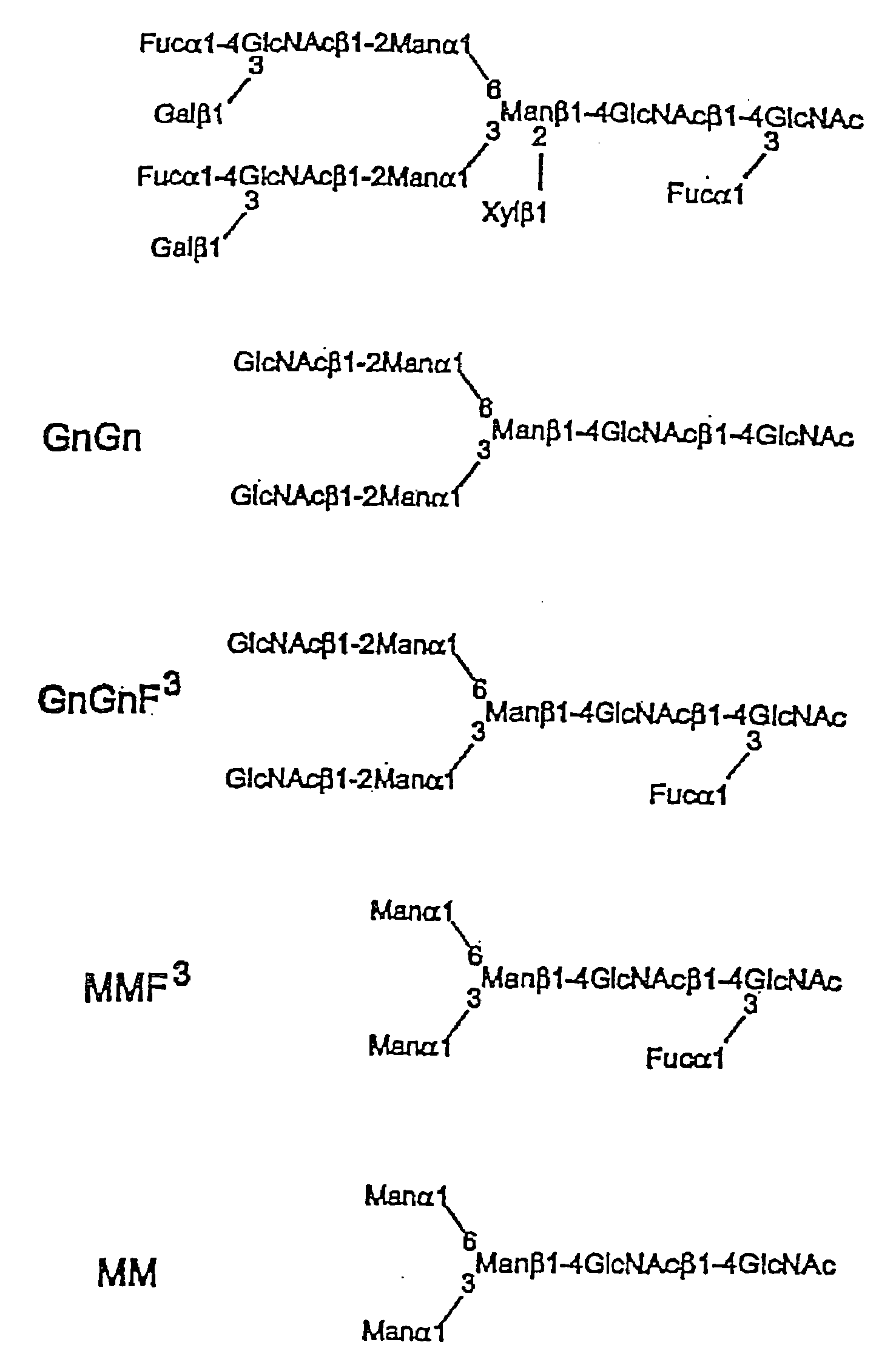
Fucosyl transferase gene
a technology of fucosyl transferase and polynucleotide coding, which is applied in the direction of transferases, peptide/protein ingredients, immunological disorders, etc., can solve the problems of plant protein induced immunological reactions in the human body
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0093]Isolation of the Core-α1,3-Fucosyl Transferase
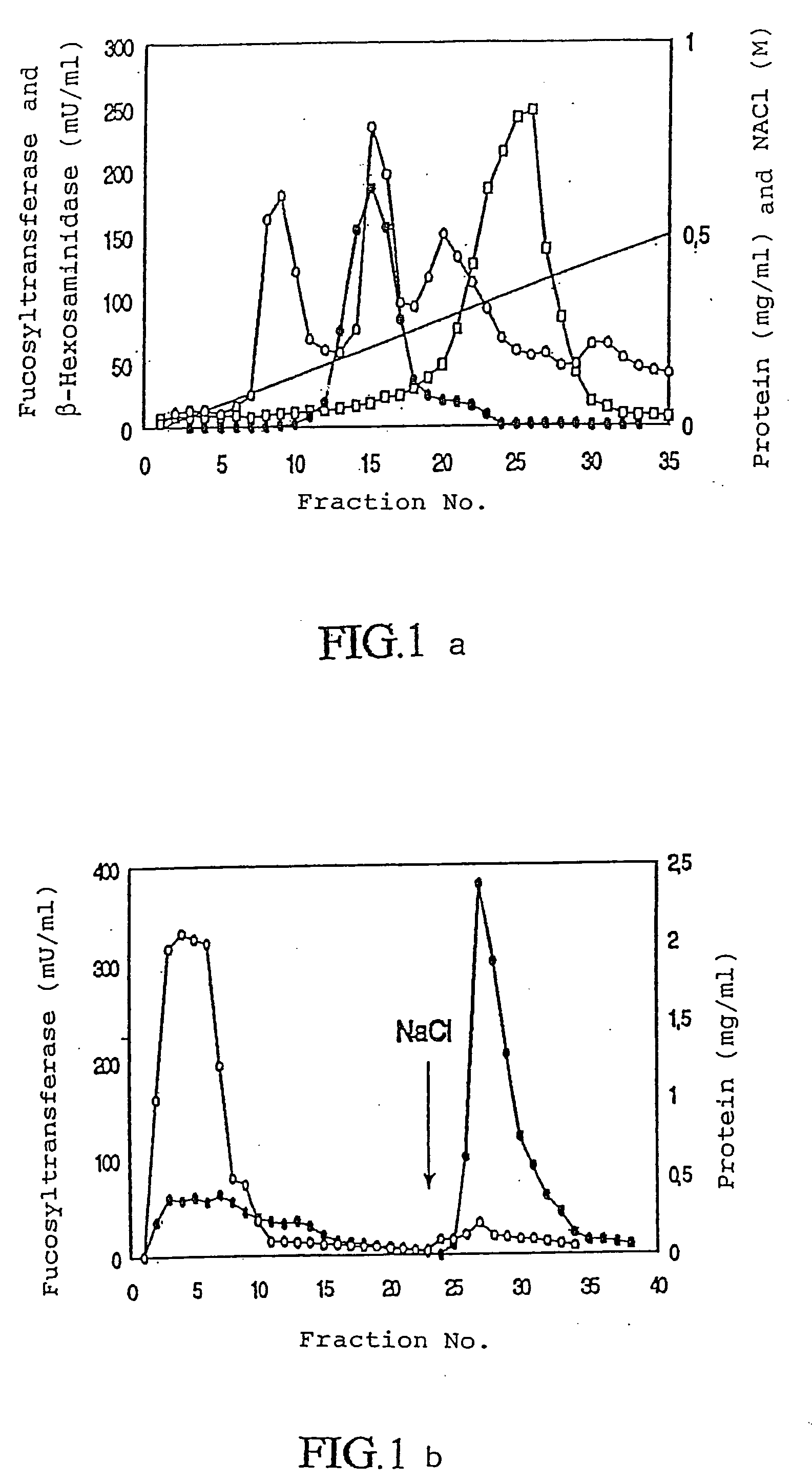
[0094]All the steps were carried out at 4° C. Mung bean seedlings were homogenized in a mixer, 0.75 volumes of extraction buffer being used per kg of beans. Subsequently, the homogenate was filtered through two layers of cotton fabric, and the filtrate was centrifuged for 40 min at 30000×g. The supernatant was discarded, and the pellet was extracted with solution buffer over night with continuous stirring. Subsequent centrifugation at 30000×g for 40 min yielded the triton extract.
[0095]The triton extract was purified as follows:
[0096]Step 1: The triton extract was applied to a microgranular diethyl amino ethyl cellulose anion exchanger DE52 cellulose column (5×28 cm) from Whatman, which previously had been calibrated with buffer A. The non-bound fraction was further treated in step 2.
[0097]Step 2: The sample was applied to an Affi-Gel Blue column (2, 5×32) column calibrated with buffer A. After washing of the column with this buffe...
example 2
[0127]SDS-PAGE and Isoelectric Focussing
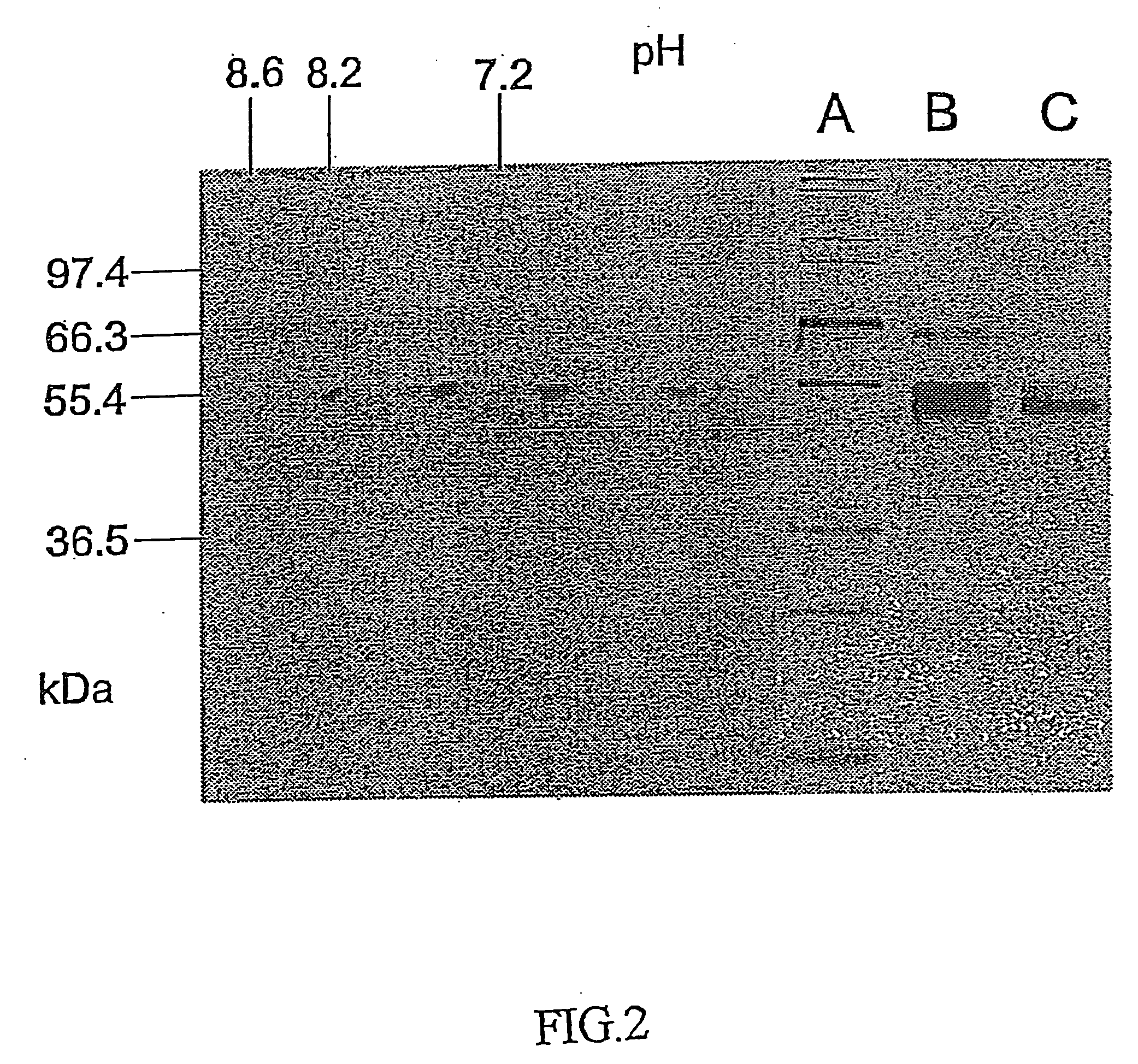
[0128]An SDS-PAGE was carried out in a Biorad Mini-protean cell on gels with 12.5% acrylamide and 1% bisacrylamide. The gels were stained either with Coomassie Brilliant Blue R-250 or Silver. Isoelectric focussing of the fucosyl transferase was carried out on prefabricated gels having a pI range of between 6-9 (Servalyt precotes 6-9, Serva). The gels were stained with silver according to the producer's protocol. For the two-dimensional electrophoresis, lanes were cut out of the focussing gel, treated with S-alkylating reagents and SDS and subjected to an SDS-PAGE, as described above.
[0129]FIG. 2 shows the illustration of an electrophoresis gel of GlcNAc-α1,3-fucosyl transferase, the two-dimensional electrophoresis being indicated on the left-hand side, and the one-dimensional SDS-PAGE being illustrated on the right-hand side. The lane denoted by A is a standard, the lane denoted by B is the GlcNAc-α1,3-fucosyl transferase from the GnGn-Sepharo...
example 3
[0131]Peptide Sequencing
[0132]For sequencing of the protein, bands were cut out of the Coomassie-stained SDS-Polyacrylamide gel, carboxyamidomethylated and cleaved with trypsin according to Görg et al. 1988, Electrophoresis, 9, 681-692. The tryptic peptides were separated with the reverse phase HPLC on a 1.0×250 mm Vydac C18 at 40° C. at a flow rate of 0.05 ml / min, wherein a HP 1100 apparatus (Hewlett-Packard) was used. The isolated peptides were separated with a Hewlett-Packard G1005 A Protein Sequencing System according to the producer's protocol. Furthermore, the peptide mixture was analyzed by Ingel digestion with MALDI-TOF MS (see below).
[0133]FIG. 4 shows the N-terminal sequences of 4 tryptic peptides 1-4 (SEQ ID NO: 5-8). Departing from the first three peptides, primers S1, A2 and A3 were prepared (SEQ ID NO: 9-11).
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com