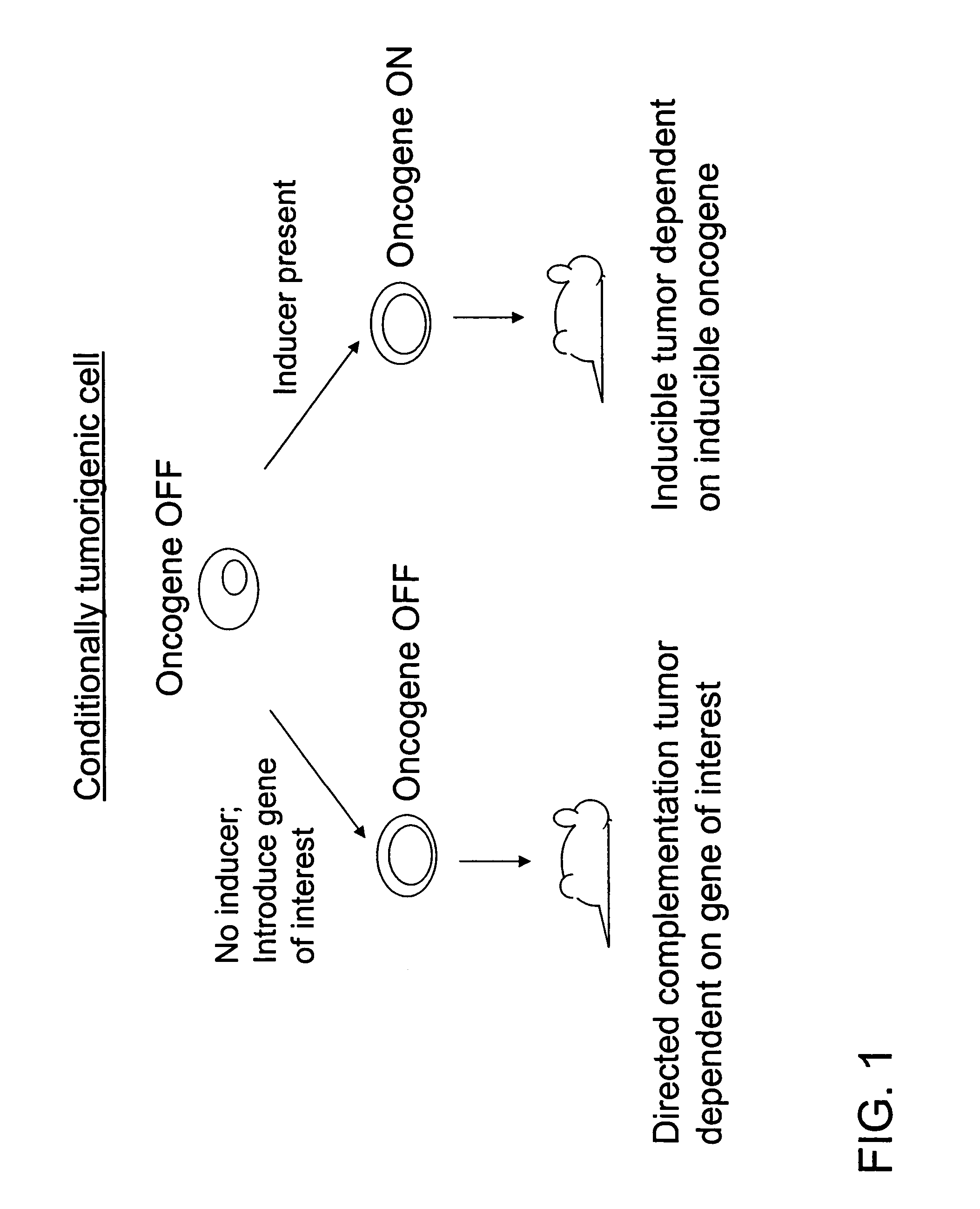
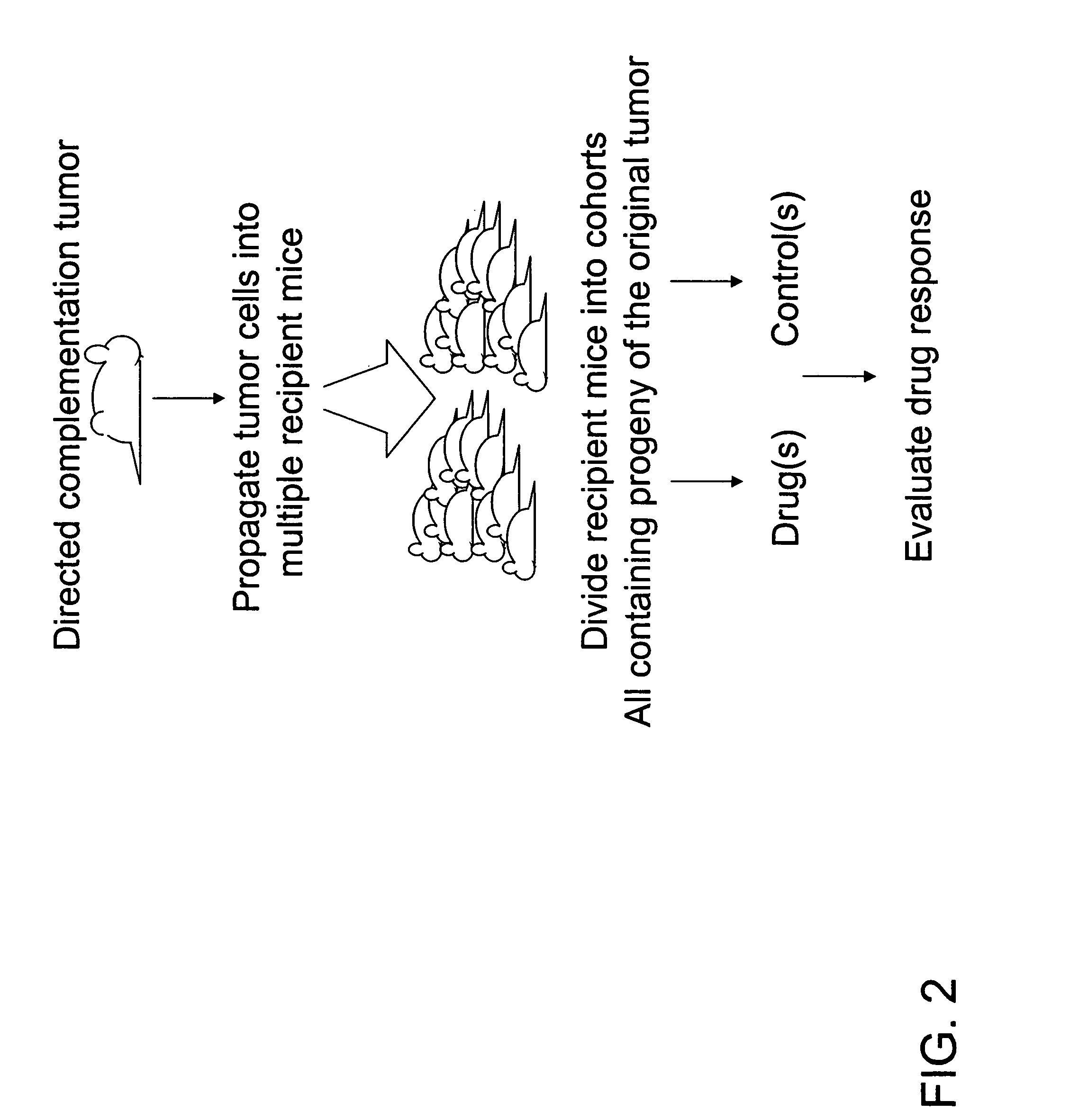
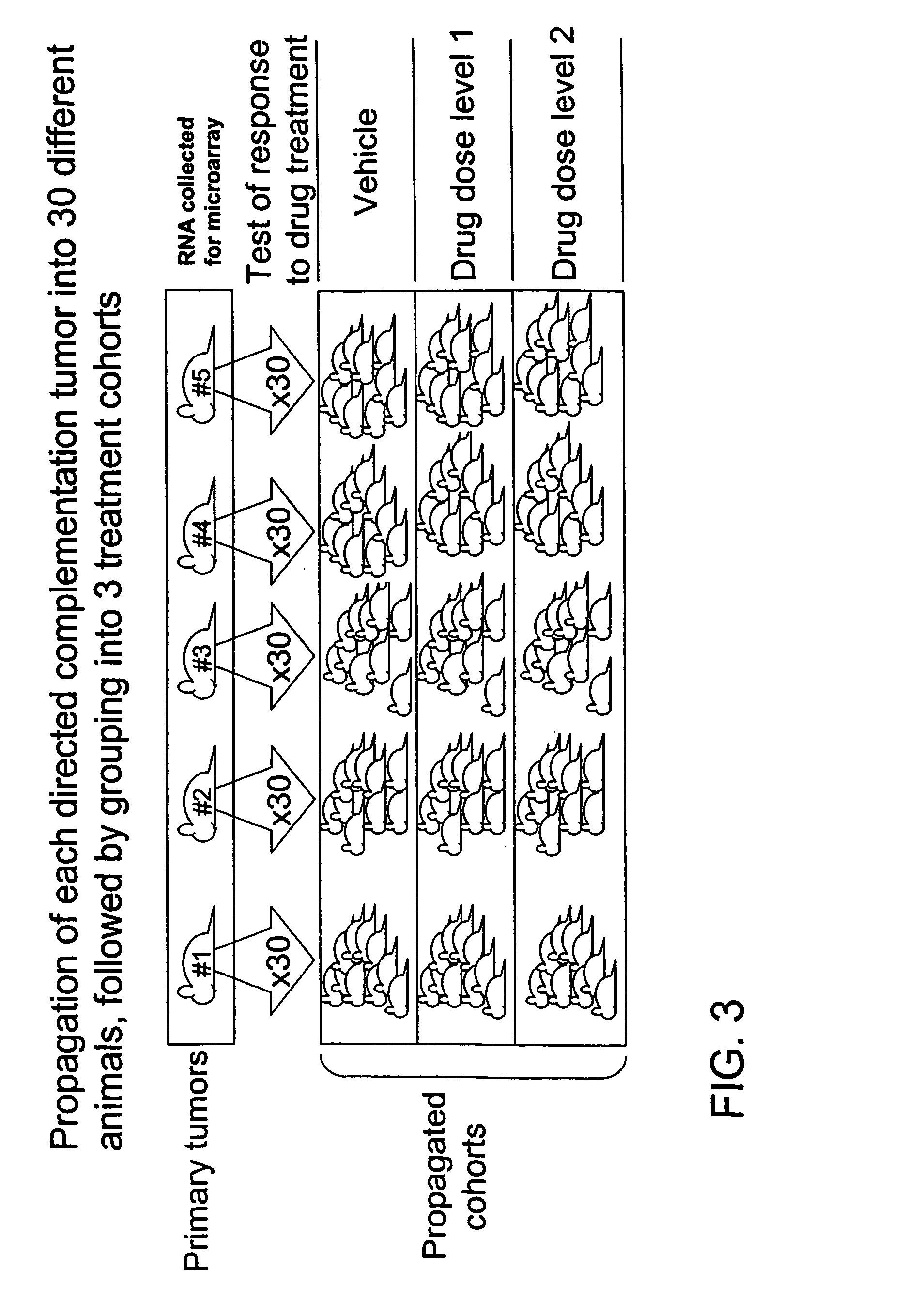
Directed complementation
a technology of complementation and directed complementation, which is applied in the field of molecular biology, oncology and drug development, can solve the problems of impracticality in designing preclinical drug development studies around and the likely incomplete picture of human cancer cell lines
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Spontaneous Inducible Tumors
[0059] Chimeric mice were engineered to develop Her2-dependent, inducible spontaneous breast tumors for use as a source of conditionally tumorigenic cells. The mice were produced as follows.
[0060] Ink4a homozygous null ES cells were co-transfected with the following four constructs, as separate fragments: MMTV-rtTA, TetO-Her2V664Eneu, TetO-luciferase and PGK-puromycin. Puromycin-resistant cells were genotyped by PCR and Southern blot. Inducibility of the oncogenes in ES cells was analyzed by northern blot. The transfected ES cells were injected into C57BL / 6 blastocysts, which were transplanted into pseudo-pregnant female mice for gestation leading to birth of the chimeric mice.
[0061] The mouse mammary tumor virus long terminal repeat (MMTV) was used to drive breast-specific expression of the reverse tetracycline transactivator (rtTA). The rtTA provided for breast-specific expression of the HER2 activated oncogene when doxycycline was provided to the mi...
example 2
Tumor Propagation
[0066] Following induction of the tetracycline-responsive promoter by doxycycline, the mice developed mammary tumors with a latency of about 2-4 months. Approximately five primary breast tumors were size-selected (>0.4 g) from one animal (two from thoracic and three from inguinal glands) so as to provide enough tumor cells for propagation.
[0067] About 0.2 g of surgically resected primary tumours were minced and digested with 1.5 ml of digestion mix (4 mg / ml collagenase; Calbiochem cat. No. 234153) in RPMI media (Gibco cat. No. 11875-093) in a 10 ml polypropylene tube for about 2 hours at 37° C. with constant shaking at 400 rpm. The digested tissues were triturated by passing them through 18 gauge needle or using cell strainers (40 μm (BD Falcon, cat. No. 352340) and 100 μm (BD Falcon cat. No. 352360) filters). The resultant cell mix was let stand at room temperature for about five minutes, followed by aspiration of only the top portion without disturbing the undig...
example 3
[0069] When the tumors reached approximately 5 mm3 in size, the treatments were begun. The animals were given three doses with 5-day intervals. The experiment was terminated one week after the final dosing. Among propagated tumors derived from any given primary tumor, uniformity in terms of latency, growth rate and drug response was expected (intra-tumoral uniformity). In contrast, among propagated tumors derived from different primary tumors variation in terms of growth rate, latency and drug response was expected (inter-tumoral variation).
[0070] As expected, intra-tumoral uniformity and inter-tumoral variation in the latency and drug response from the propagated tumors was observed. It was scored as a positive response if a tumor showed a statistically significant difference in size between vehicle and drug treatment at the end of one week after last dosing. Propagated tumors that came from only one of the five primary tumors showed significant reduction in tu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com