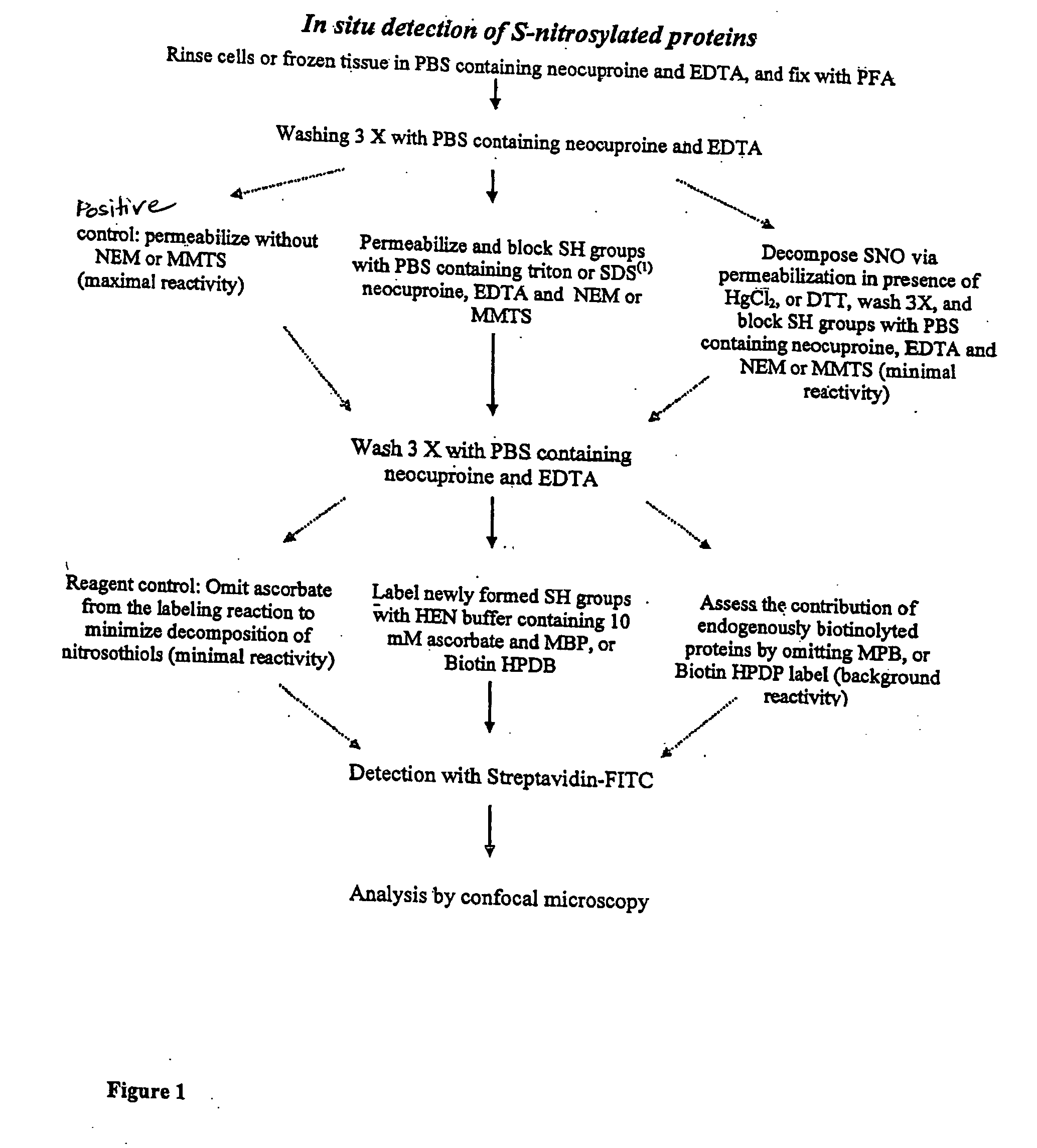
Detection of nitrosylated proteins
a technology of nitrosylated proteins and detection methods, applied in the direction of electrolysis, isotope separation, enzyme stabilisation, etc., can solve the problem of limitations in the methodology typically used
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0047] This example describes certain protocols and methods that may be useful in various embodiments of the invention.
[0048] A line of spontaneously transformed mouse lung alveolar type II epithelial cells (C10) was used in some experiments, as described below. C10 cells were cultured in CMRL1066 medium containing 10% fetal bovine serum (FBS, GIBCO). In experiments involving microscopic analysis, the cells were grown on glass cover slips. At about one hour prior to exposure to a test agent, the medium was switched to phenol red free DMEM / F12 containing 0.5% FBS. Some cells were exposed to S-nitroso-glutathione (GSNO) or S-nitroso-N-acetyl penicillamine (SNAP) for about 1 hour, in the presence of an equimolar concentration of freshly prepared L-cysteine. To monitor S-nitrosylation in response to NOS2 activation, the cells were treated with 10 ng / ml TNF alpha (TNFα) and 100 U / ml interferon gamma (IFNγ), for about 24 hours. Incubation with L-N-monomethyl arginine (L-NMMA) for about 1...
example 2
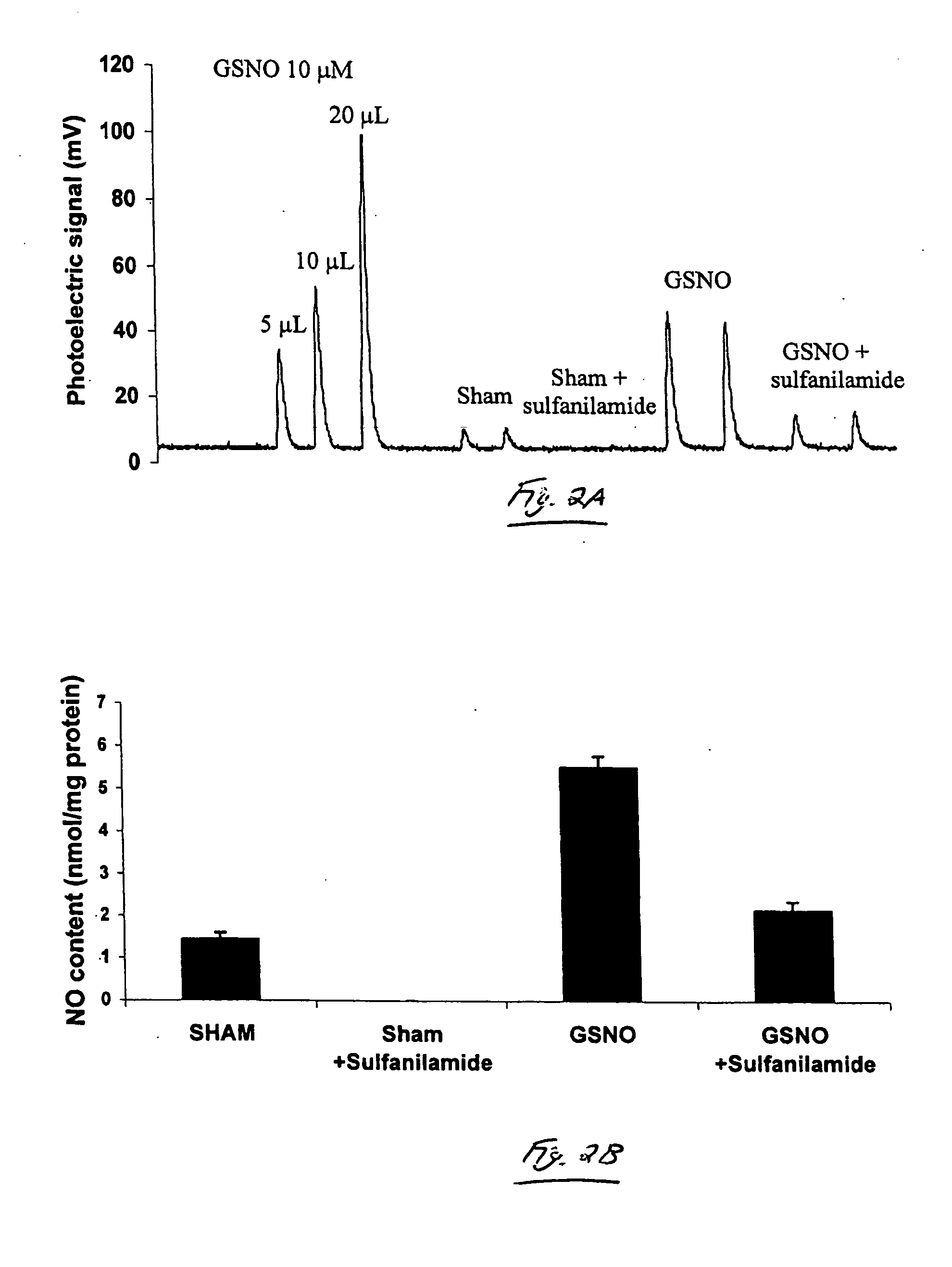
[0054] This example demonstrates NO detection by chemiluminescence, using one embodiment of the invention. Using methods similar to those described in Example 1, this example demonstrates that the treatment of cells with exogenous S-nitrosothiols resulted in an increase in the SNO (nitrosothiol) content of the cells.
[0055] Mouse lung epithelial (C10) cells were incubated with 1 mM GSNO for about one hour, lysed, and the NO released following reduction in acidic KI / I2 was detected by chemiluminescence. As shown in FIG. 2, the total NO content in lysates from cells treated with GSNO was found to have increased, as compared with lysates from non-treated cells. In order to eliminate the contribution of nitrites to the observed photoelectric signal, the cell lysates were incubated with a nitrite quencher, sulfanilamide, and assessed the sulfanilamide resistant NO signal by chemiluminescence. Under these experimental conditions, employing 12 micrograms of protein, S-nitrosthiol was not d...
example 3
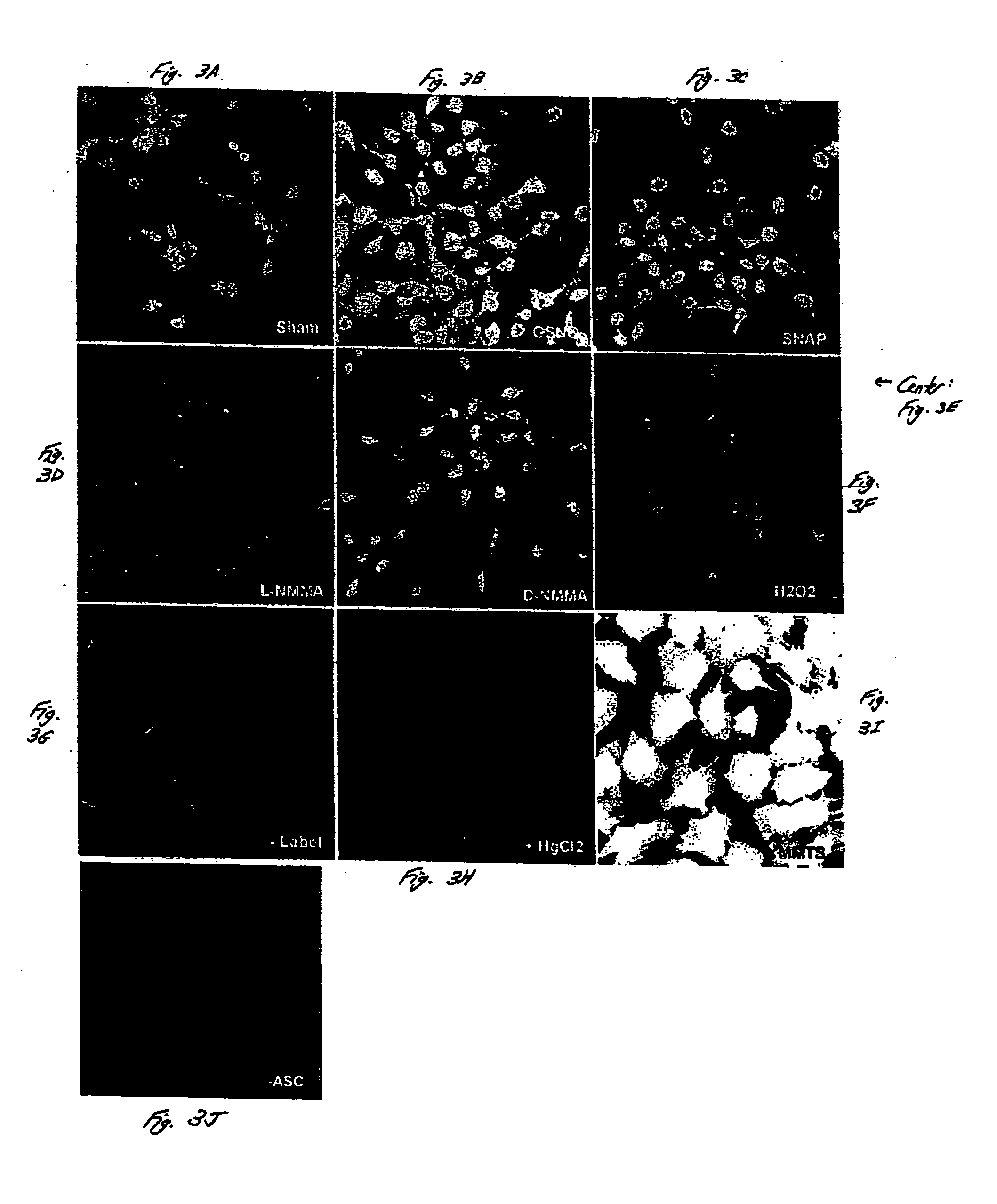
[0057] The detection of S-nitrosylated proteins in situ is demonstrated in this example, in accordance with an embodiment of the invention. Intact C10 cells were studied using methods similar to those described in Example 1. As shown in FIG. 3, S-nitrosylated proteins were detected in intact C10 cells using biotinylation, streptavidin-FITC and analysis by confocal laser scanning microscopy. C10 cells were exposed to 1 mM SNAP (FIG. 3C) or 1 mM GSNO (FIG. 3B) for about 1 hour, or to L-NMMA (FIG. 3D) or D-NMMA (FIG. 3E) for about 16 hours, fixed, and subjected to biotin derivatization, incubation with streptavidin-FITC, and confocal microscopy.
[0058] In non-treated cells (sham controls, FIG. 3A), some endogenous reactivity was observed. This reactivity was inhibited upon the addition of HgCl2, which decomposes SNO prior to blocking with methyl methanethiosulfonate (MMTS) (FIG. 3H). The omission of ascorbate, which reduces SNO bonds prior to the addition of the biotin-HPDP label, also...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com