Carboxylesterase inhibitors
a technology of carboxylesterase and inhibitors, applied in the field of carboxylesterase inhibitors, can solve the problems of delayed diarrhea, high toxicity of compounds, cell death, etc., and achieve the effect of increasing the effectiveness of insecticides and pesticides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Carboxylesterase Inhibition Assay
(a) ortho-nitrophenyl Acetate (o-NPA) as the Substrate
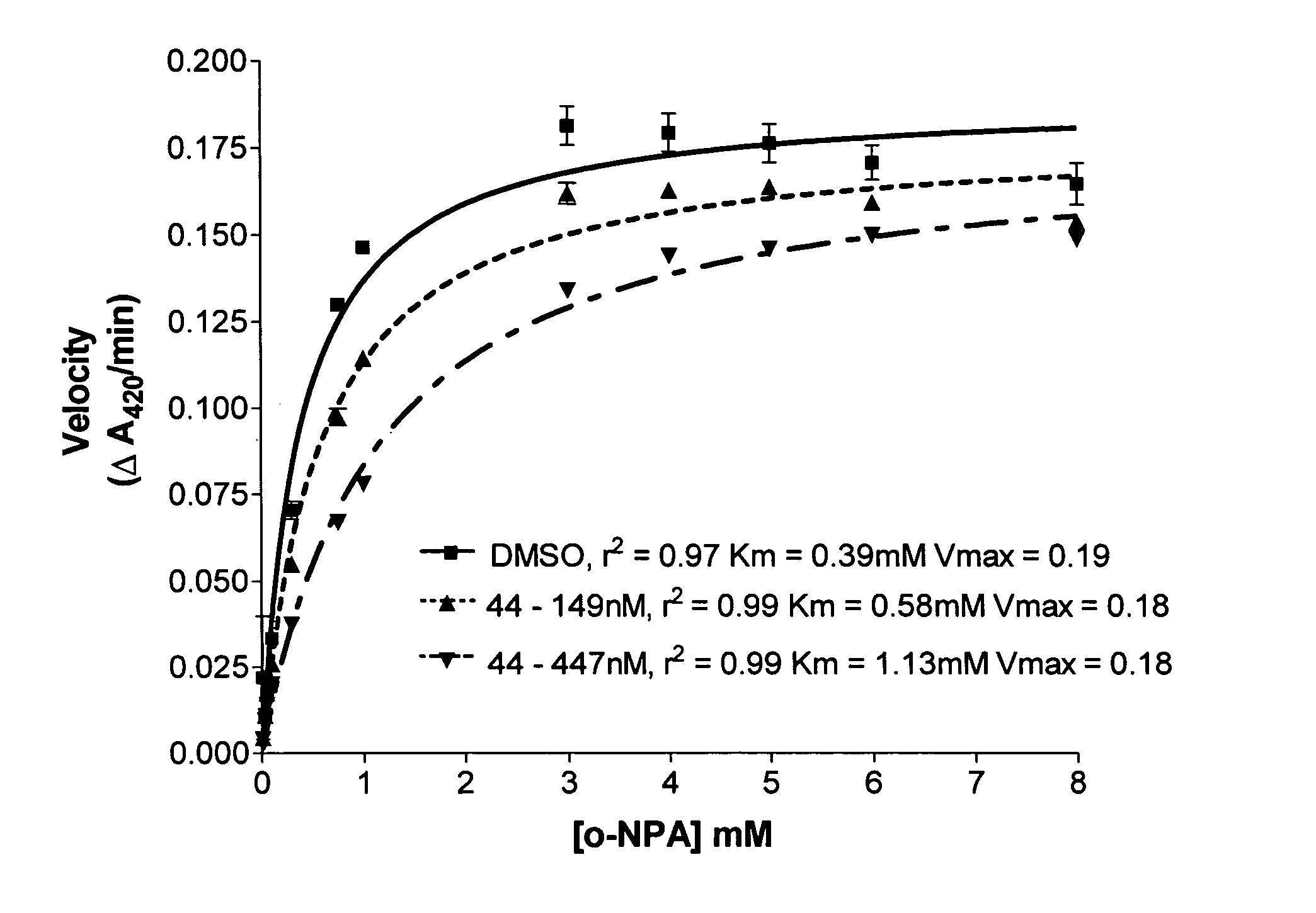
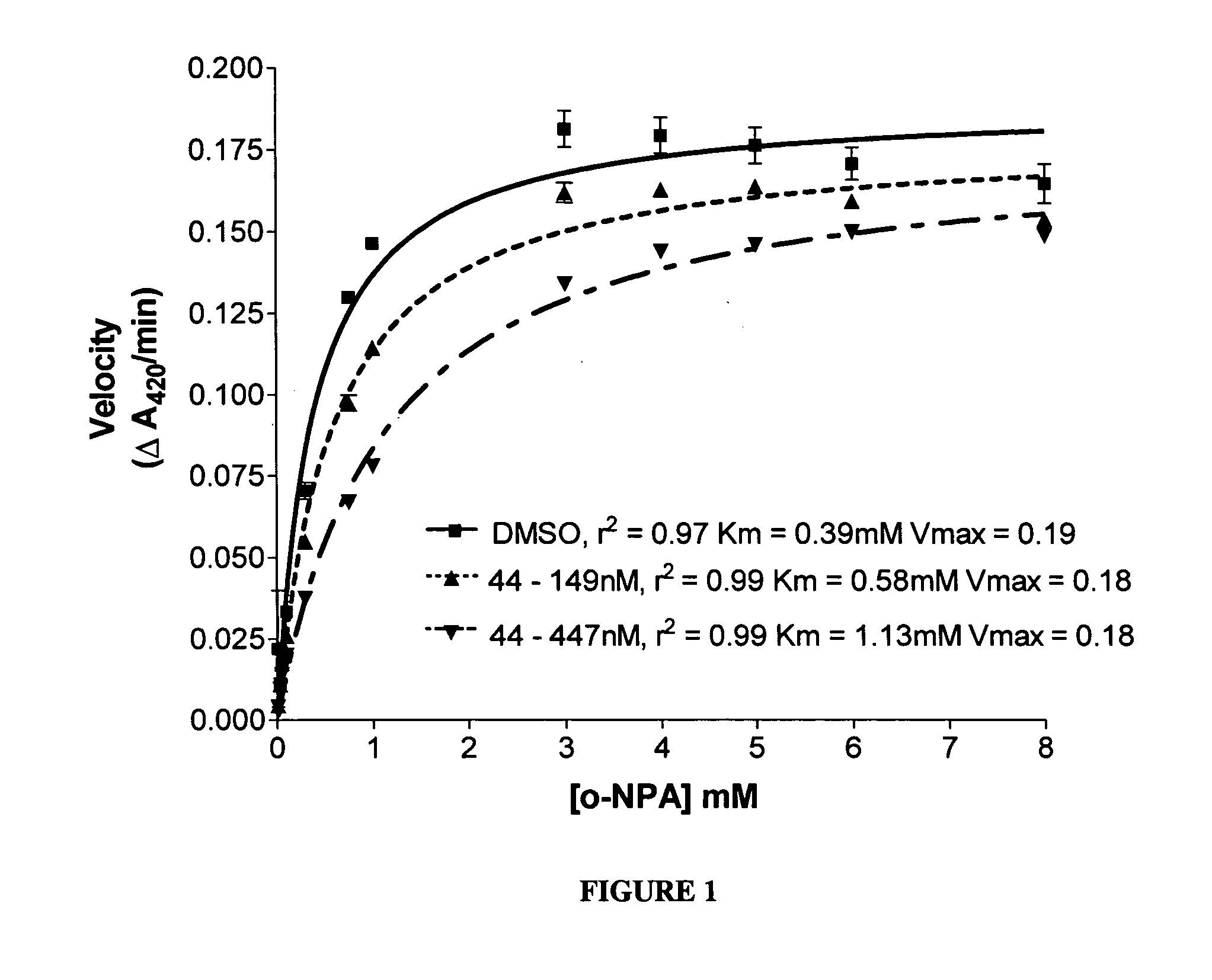
Carboxylesterase (CE) inhibition was determined by a spectroscopic assay using o-nitrophenyl acetate (o-NPA) as a substrate. Recombinant CEs produced from expression in Spodoptera frugiperda Sf9 insect cells via baculovirus were purified from serum-free culture media for the enzyme inhibition studies. Enzymes were incubated in 200 μl of 50 mM Hepes (pH 7.4) containing 3 mM o-NPA. Conversion of the o-NPA to nitrophenol was monitored by measuring the change in the absorbance at 420 nm. Absorbance readings were taken every 15 seconds for 2 minutes, and data transferred to a computer data spreadsheet. Inhibitors were dissolved in dimethylsulfoxide (DMSO) and inhibition of CE activity determined by the addition of the inhibitor solution to the o-NPA reaction mixture at a concentration of 100 μM. The final DMSO concentration was always 1% or less. The Ki value (i.e., the concentration of inhibitor t...
example 2
Inhibition of Acetylcholinesterase and Butyrylcholinesterase
The activity of the hiCE selective carboxylesterase inhibitors identified in Table 3 toward acetylcholinesterase (AcChE) and butyrylcholinesterase metabolism was investigated. Purified AcChE was purchased from Sigma Biochemicals (St. Louis, Mo.) and substrate metabolism was monitored using a spectrophotometric assay. 1 mM acetylthiocholine was mixed with 0.5 mM of 5,5′-dithio-bis-(2-nitrobenzoic acid) in 50 mM Hepes (pH 7.4) in the presence of the inhibitor (100 μM). The reaction was initiated by the addition of 0.22 U / ml AcChE (where 1 U is the amount of enzyme that hydrolyzes 1 μmol of acetylthiocholine iodide per min at pH 7.4 at 37° C.), and the reaction monitored by measuring the change of the absorbance at 405 nM every 15 seconds 2 minutes. Data was transferred to GraphPad Prism software, and Ki values were calculated as described in the Example above.
Purified BuChE was purchased from Sigma Biochemicals (St. Louis...
example 3
Ability of the Selective hiCE Inhibitors to Cross Cell Membranes
To monitor the ability of the compounds to cross the cell membrane and inhibit carboxylesterase activity intracellularly, an assay using human tumor cells expressing hiCE was devised. U373MGhiCE (a human glioma cell line transfected with a plasmid expressing hiCE) were plated in T25 flasks and allowed to grow to confluency (approximately 2-5×105 cells). Inhibitors were then added at a final concentration of 10 μm, and incubation allowed to continue for 1 hour. At this time, the cells were washed extensively with 5 ml of complete media and 2×5 ml of Hanks balanced salt solution, and cell extracts were prepared. Carboxylesterase activity was then determined using o-NPA as a substrate.
Table 8 sets forth the level of intracellular enzyme inhibition following incubation of human glioblastoma tumor cells expressing human intestinal carboxylesterase with 10 μM of each compound identified in Table 3.
TABLE 8Inhibition of i...
PUM
| Property | Measurement | Unit |
|---|---|---|
| crystal structure | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| structure | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com