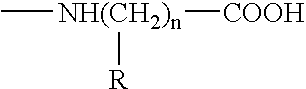Hepatitis B virus binding proteins and uses thereof
a technology binding proteins, which is applied in the field of hepatitis b virus binding proteins, can solve the problems of high risk of liver cirrhosis and primary liver carcinoma, no antiviral drugs with good and long-term efficacy, and no molecules of these molecules have been convincingly tied
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials and Methods
[0143] Preparation of the affinity columns: For the preparation of the affinity columns first a recombinant preS1 protein was prepared. The HBV preS1 gene was obtained by PCR amplification from a plasmid containing the entire HBV genome (cloned at the laboratory of W. J. Rutter at the UCSF). The following primers were used for amplification: 5'-GGAGATCTTCAAAACCTGGCAAAGGC-3' (SEQ ID NO:10) and 5'-GAATTCCACTGCATGGCCTG-3' (SEQ ID NO:11). The PCR product was cloned into the p-RSET-B vector (Invitrogene). The constructed plasmid was sequenced using the Weizmann Institute service center (see, SEQ ID NO:7). Recombinant His tagged pre-SI protein (see SEQ ID NO:8) was expressed in E. coli B121 cells. Cells were grown overnight at 37.degree. C. in M9ZB medium containing 0.4% glucose. The overnight culture was then diluted 1:50 with fresh M9ZB medium and was further grown at 37.degree. C. When the OD(600 nm) reached 0.7-0.8 the cells were induced with IPTG (1 mM). The solu...
example 2
Production of the Recombinant HBV preS1 Protein
[0154] Based on previous research, the preS1 region of HBsAg is expected to contain the receptor binding region (Neurath et al., 1985; Petit et al., 1991). For HBV receptor purification a recombinant His-tagged preS1 protein (FIGS. 1a and 1b, SEQ ID NOs:7-8) was prepared. The recombinant protein was purified to homogeneity by employing a Ninta-affinity column (Quiagene). Also, a 29 amino-acid long peptide that was reported to be sufficient to interact with hepatocytes was synthesized (SEQ ID NO:9). This synthetic peptide was further purified on a G25 column to obtain a homogenous peptide (FIG. 1c).
example 3
Purification of HBV preS1-Binding Proteins
[0155] The recombinant preS1 protein (SEQ ID NO:7) was covalently cross-linked to beads (Affinity gel 10, BioRad) according to the manufacturer's instructions and was used for affinity chromatography. Concentrated urine (X 1000) was passed through the column, the column was washed and the bound proteins were eluted at low pH (see methods). The eluted fractions were analyzed on SDS-PAGE gel and silver stained. Two major bands appeared after elution from the preS1 column (FIG. 2, lane E2). The estimated molecular masses of the stained proteins were 50 and 43, and therefore they were named UP50 and UP43, respectively.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| melting temperatures | aaaaa | aaaaa |
| melting temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com