Chip copying method for DNA computation
A replication method and DNA probe technology, applied in the field of DNA chips, can solve the problem of only replicating a few chips, and achieve the effect of large-scale mechanization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
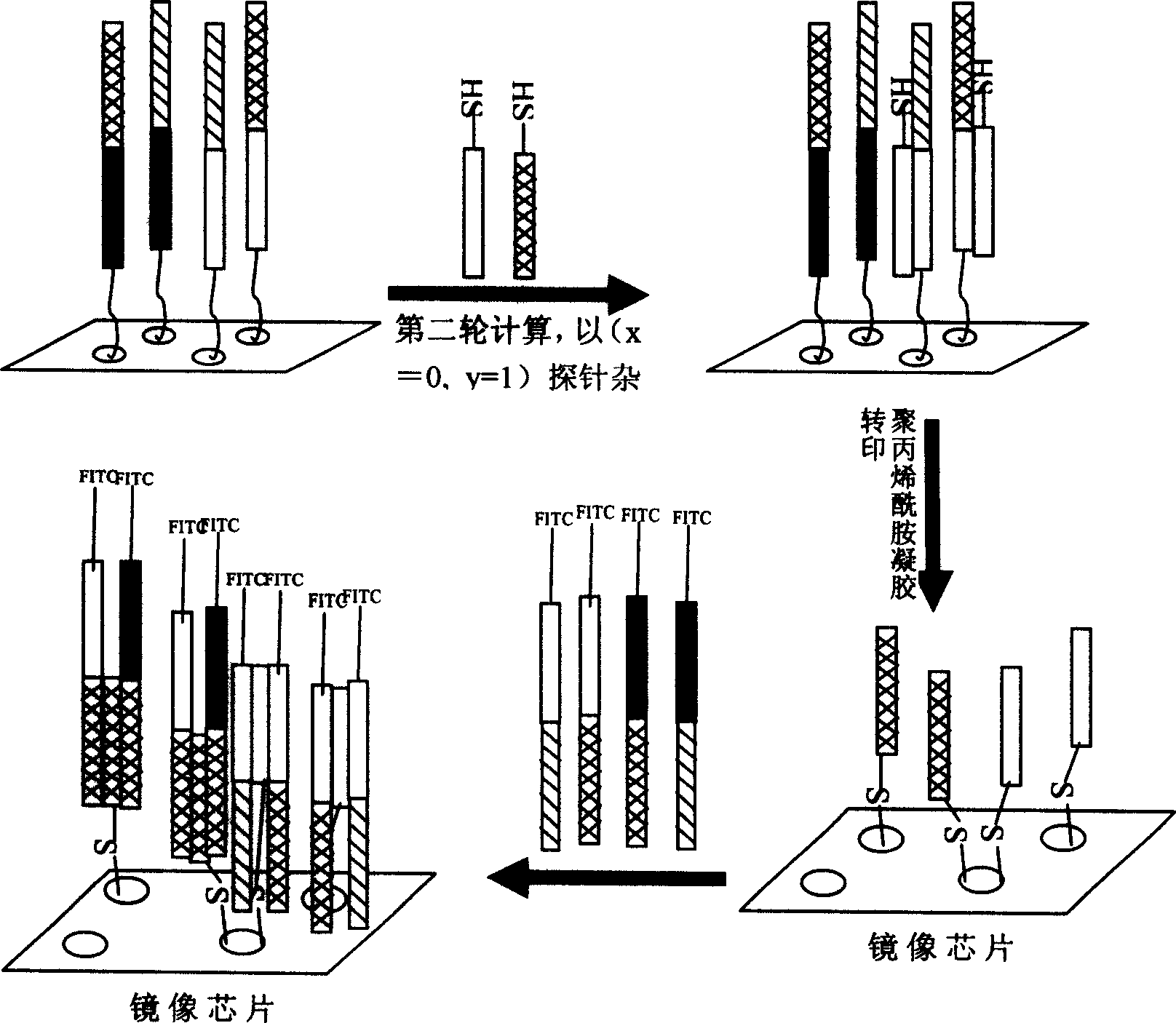
[0012] Embodiment 1, based on the unrestricted replication of the DNA chip of thiol and polyacrylamide gel system
[0013] The first step is to prepare three sets of DNA materials A, B, and C. The A set is used to construct the mother chip. The sequence and length of the C set are the same as those of the A set except for the sulfhydryl label at the end. The sequence and length of the B set are the same as those of the A set. The materials in group A and C are complementary, and they are complementary probes for the materials in group A and C, and the ends of the DNA in group B are labeled with sulfhydryl groups.
[0014] In the second step, the mother chip is constructed with group A DNA materials.
[0015] The third step is to use group B probes to hybridize with the mother chip at 25-50° C. for half an hour, and then wash.
[0016] The fourth step is to pour the polymerization reaction solution system of acrylamide monomer and its crosslinking agent on the mother chip area...
Embodiment 2
[0021] Embodiment 2, based on the unrestricted replication of the DNA chip of thiol and polyacrylamide gel system
[0022] The first step is to prepare three sets of DNA materials A, B, and C. The A set is used to construct the mother chip. The sequence and length of the C set are the same as those of the A set except for the sulfhydryl label at the end. The sequence and length of the B set are the same as those of the A set. The materials in group A and C are complementary, and they are complementary probes for the materials in group A and C, and the ends of the DNA in group B are labeled with sulfhydryl groups.
[0023] In the second step, the mother chip is constructed with group A DNA materials.
[0024] The third step is to use group B probes to hybridize with the mother chip at 25-50° C. for half an hour, and then wash.
[0025] The fourth step is to pour the polymerization reaction solution system of acrylamide monomer and its crosslinking agent on the mother chip area...
Embodiment 3
[0030] Embodiment 3, based on the unrestricted replication of the DNA chip of thiol and polyacrylamide gel system
[0031] The first step is to prepare three sets of DNA materials A, B, and C. The A set is used to construct the mother chip. The sequence and length of the C set are the same as those of the A set except for the sulfhydryl label at the end. The sequence and length of the B set are the same as those of the A set. The materials in group A and C are complementary, and they are complementary probes for the materials in group A and C, and the ends of the DNA in group B are labeled with sulfhydryl groups.
[0032] In the second step, the mother chip is constructed with group A DNA materials.
[0033] The third step is to use group B probes to hybridize with the mother chip at 25-50° C. for half an hour, and then wash.
[0034] The fourth step is to pour the polymerization reaction solution system of acrylamide monomer and its crosslinking agent on the mother chip area...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com