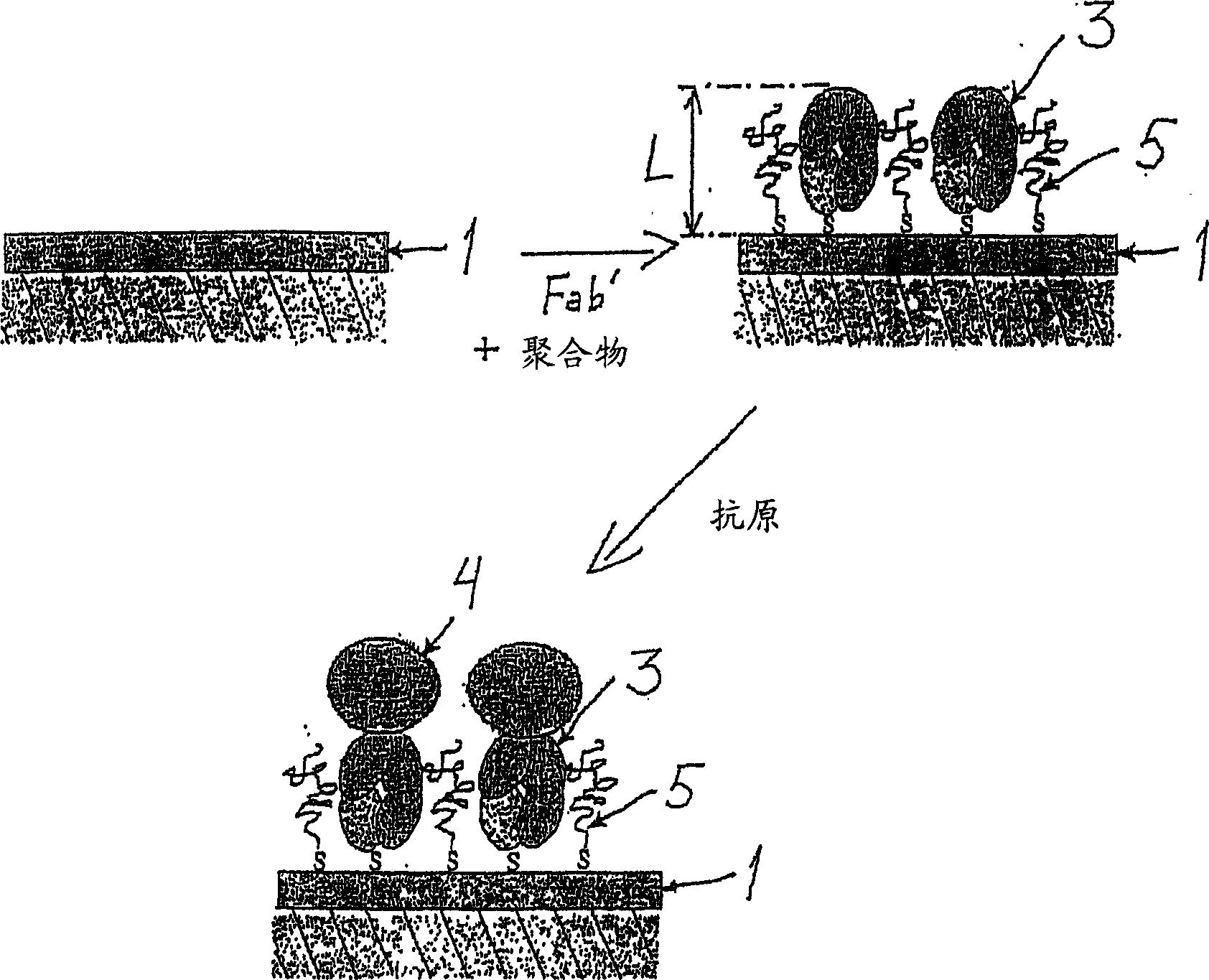
Diagnostic methods
An antigen, Helicobacter pylori technology, applied in the field of diagnosis of Helicobacter pylori infection, can solve the problems of infection risk, dislike of handling stool samples, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] General procedure for preparing the carrier substrate of the invention
[0044] To prepare the carrier substrate, Fab' fragments were first prepared from specific monoclonal anti-H. pylori antibodies as follows. First, use ImmunoPure F(ab′) 2 Preparation kit (PIERCE, USA) for preparation of F(ab') from monoclonal anti-H. pylori antibodies such as monoclonal anti-H. 2 fragment. Other known commercially available kits and methods may likewise be used. F(ab') was then dithiothreitol (DTT, Merck) in HEPES / EDTA buffer 2 Fragments were split into Fab' fragments with HEPES / EDTA buffer containing 150 mM NaCl, 10 mM HEPES, 5 mM EDTA, pH 6.0 as described by Ishikawa [Ishikawa, E., J. Immunoassay 4 (1983) 209- 320] typically performed overnight in microdialysis tubing. Briefly, F(ab') at a concentration of 0.2-0.5 mg / ml 2 Fragments were mixed with HEPES / EDTA buffer and 6.25 mM DTT solution in microdialysis tubing. The dialysis tubing was immersed in 250 ml of argon-purged H...
Embodiment 2
[0048] General procedure for measuring Helicobacter pylori antigen by SPR
[0049] H. pylori antigens in biological samples can be measured using the following general SPR procedure. The surface of the carrier substrate prepared in Example 1 was rinsed with PBS (pH 7.2). Negative samples (blanks) were measured first. The negative sample used varies with the biological sample to be measured. Thus, for example, when measuring a stool sample, a negative sample is a stool sample negative for H. pylori; when measuring a urine sample, a negative sample is a urine sample from a subject without H. pylori infection; when measuring a serum sample , negative samples were serum samples obtained from subjects without H. pylori infection. Then, the surface of the carrier substrate was sequentially brought into contact with the standard solution, the positive and negative control solutions and the sample, the flow cell of the measuring device was filled with each solution to be measured f...
Embodiment 3
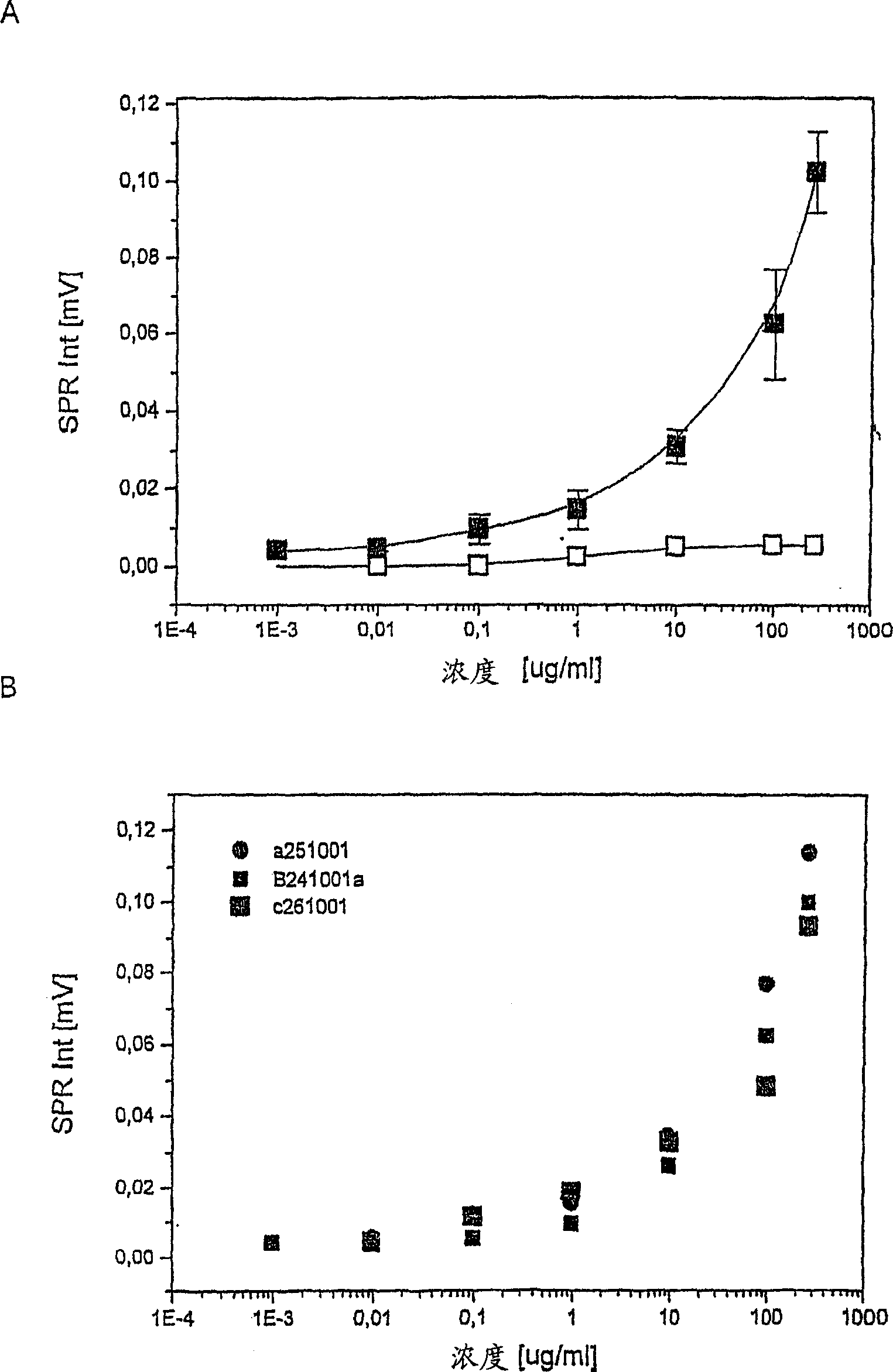
[0051] Preparation of standard curve and measurement reproducibility
[0052] H. pylori antigens used as standards were extracted from bacterial pellets of H. pylori strain ATCC 49503 using the glycine-acid extraction procedure described by Rautelin and Kosunen [J. Clin. Microbiol. 25(10) (1987) 1944-1951]. Protein concentration was determined by Bradford assay [Bradford, Anal. Biochem. 72 (1976) 248]. Standards were diluted in 0.1 M PBS (pH 7.2) to concentrations of 0.001, 0.01, 0.1, 1, 10, 100 and 270 μg / ml and measured following the general procedure described in Example 2. Three separate measurements were performed on subsequent days to analyze reproducibility.
[0053] The results are shown in Figure 3A. The standard curve shown in Figure 3A demonstrates that responses and antigen concentrations are directly comparable on a semi-log scale. Table 1 shows the standard deviation of the three standard curve measurements over subsequent days (Figure 3B). Excellent reproduc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com