Monolithic matrix for separating nucleic acids by reverse-phase into-pair high performance liquid chromatography
A reversed-phase ion pair, matrix technology, applied in the field of bio-organic molecule separation, which can solve the problems of limited efficiency, reduced surface area, and high operating pressure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Isolation of single-stranded oligonucleotides
[0062] Prepare a mixture containing 1.0 ml trimethylolpropane trimethacrylate, 0.32 ml hexyl methacrylate, 2.4 ml isooctane and approximately 20 mg azobisisobutyronitrile and inject it into an empty 4.6 mm ID× 5 cm long HPLC column tubing (one end capped with end fitting and stopper). Seal the loaded tube with an end fitting and a stopper at the other end, and then soak overnight in a water bath at 65°C. After the reaction, the stoppers and end fittings were removed, and the end fittings with polyethylene porous sheets were fitted. The entire column was flushed with tetrahydrofuran.
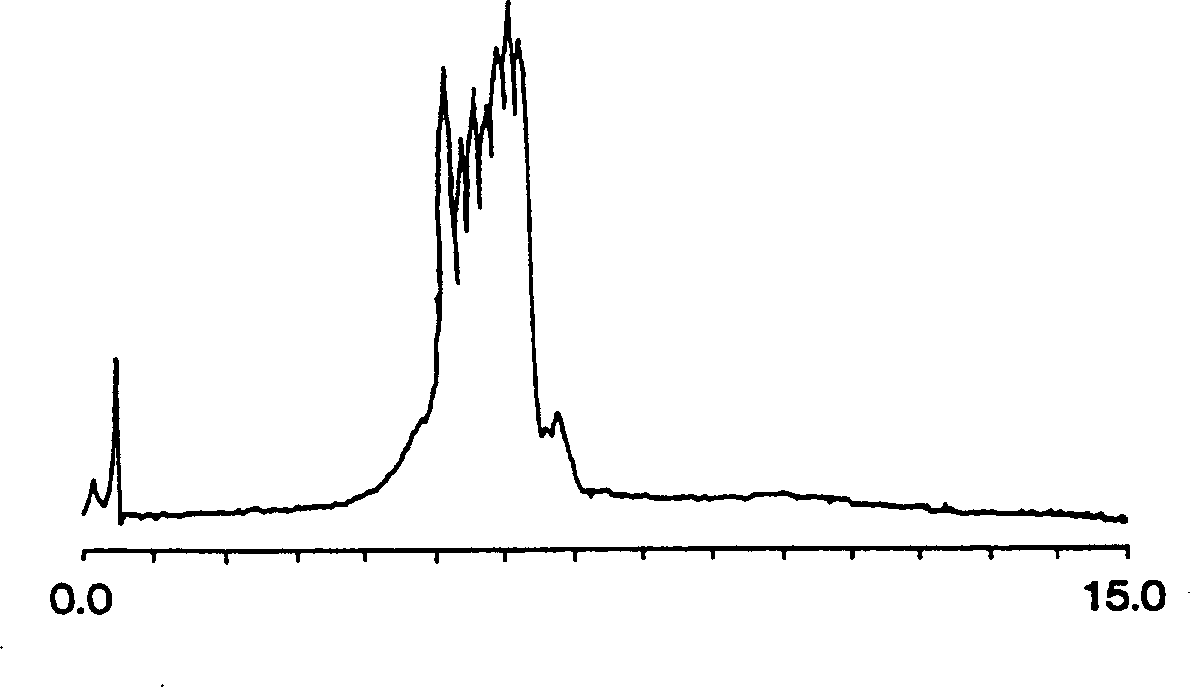
[0063] figure 1 It is a chromatogram of separating single-chain oligothymidylic acid with the C6 monolithic column of the above structure. Specifically, the sample is 10 microliters of oligothymidylates between 12-18 units in length. Oligonucleotides were eluted at a flow rate of 2 ml / min, gradient of 0-12% acetonitrile (prepared with 100 ...
Embodiment 2
[0065] Separation of dsDNA
[0066] Prepare a mixture containing 1.25 ml of trimethylolpropane trimethacrylate, 0.30 ml of lauryl methacrylate, 2.6 ml of a mixture of 95% isooctane and 5% toluene, and 17 mg of azobisisobutyronitrile , and inject it into an empty 4.6 mm ID x 5 cm long HPLC column (capped at one end with an end fitting and stopper). Seal the loaded tube with an end fitting and a stopper at the other end, and then soak overnight in a water bath at 65°C. After the reaction, the stoppers and end fittings were removed, and the end fittings with polyethylene porous sheets were fitted. The entire column was flushed with tetrahydrofuran.
[0067] Figure 2A It is a chromatogram of separating double-stranded DNA fragments with the single-chip C12 column of the above structure. Specifically, the sample was 3 microliters of pUC18 DNA (digested with MSP I restriction enzyme). DNA fragments were eluted at a flow rate of 1.0 ml / min with a 10 minute gradient of 35-60% 20...
Embodiment 3
[0072] Isolation of partially denatured double-stranded polynucleotides
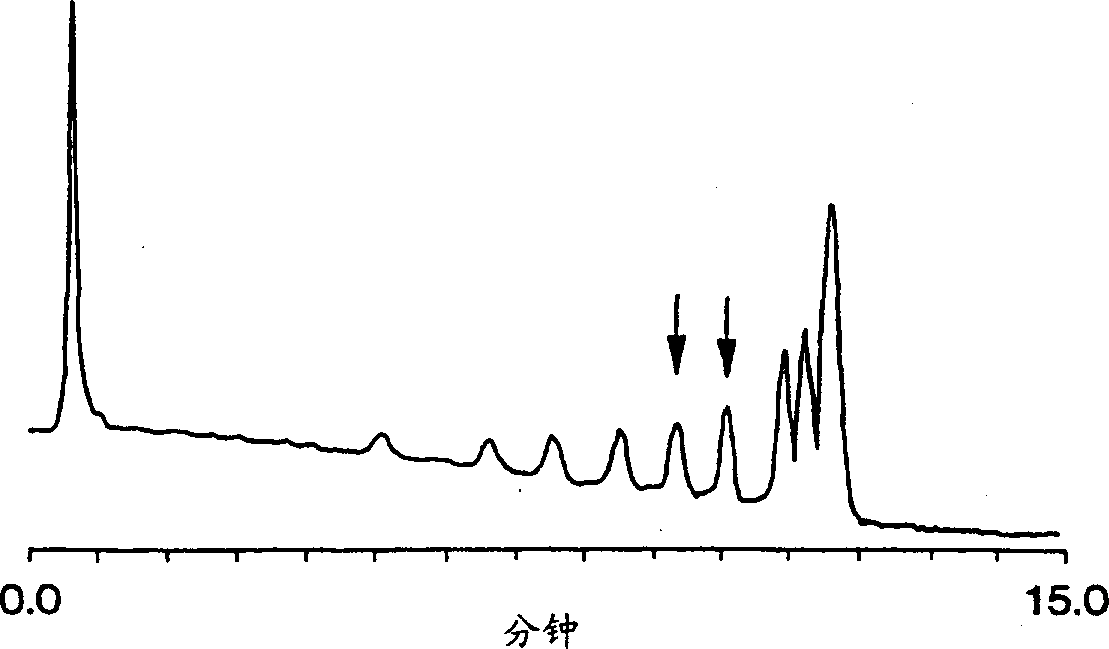
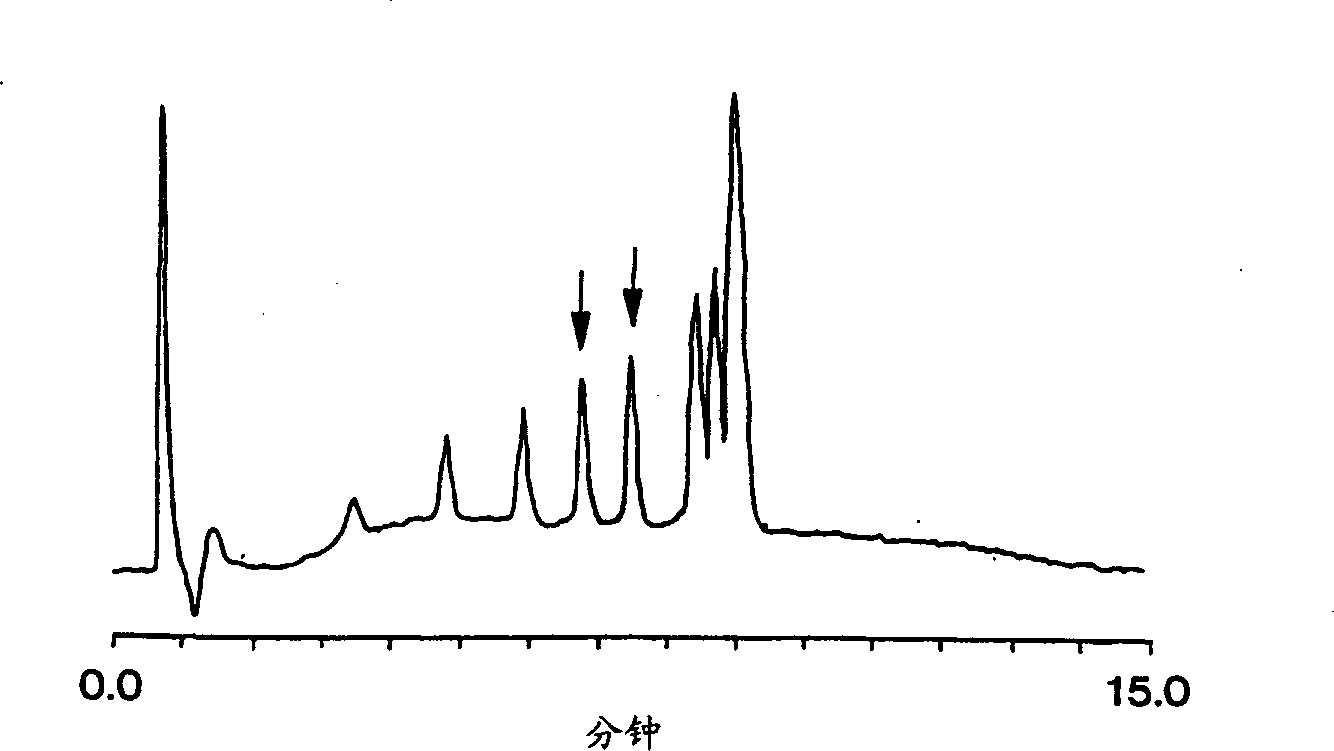
[0073] The invention can also isolate partially denatured double-stranded polynucleotides. Such as Figure 3A with 3B As shown, a porous monolithic column was used to separate a DNA fragment of 304 base pairs in length. exist Figure 3A , the sample contains one type of DNA, Figure 3B The samples in Figure 3A A mixture of the type in and a variant sequence containing a single base substitution. at a certain temperature, Figure 3B The sample shows a second peak, indicating partially denatured DNA with a single base substitution.
[0074] Specifically, in Table 3A, homogeneous double-stranded DNA with a length of 304 base pairs was separated on the monolithic 12 column prepared as above. Table 2 lists the conditions under which this separation was performed, as follows:
[0075] mobile phase:
[0076] observed under similar conditions Figure 3B data, but contains samples other than ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com