Phage-assisted cellooligosaccharide transporter continuous directed evolution system and method
A directed evolution, cello-oligosaccharide technology, applied in the field of genetic engineering, can solve the problems of reducing evolutionary efficiency, human intervention, and high cost, and achieve the effects of avoiding human intervention, high evolutionary efficiency, and improving transport capacity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
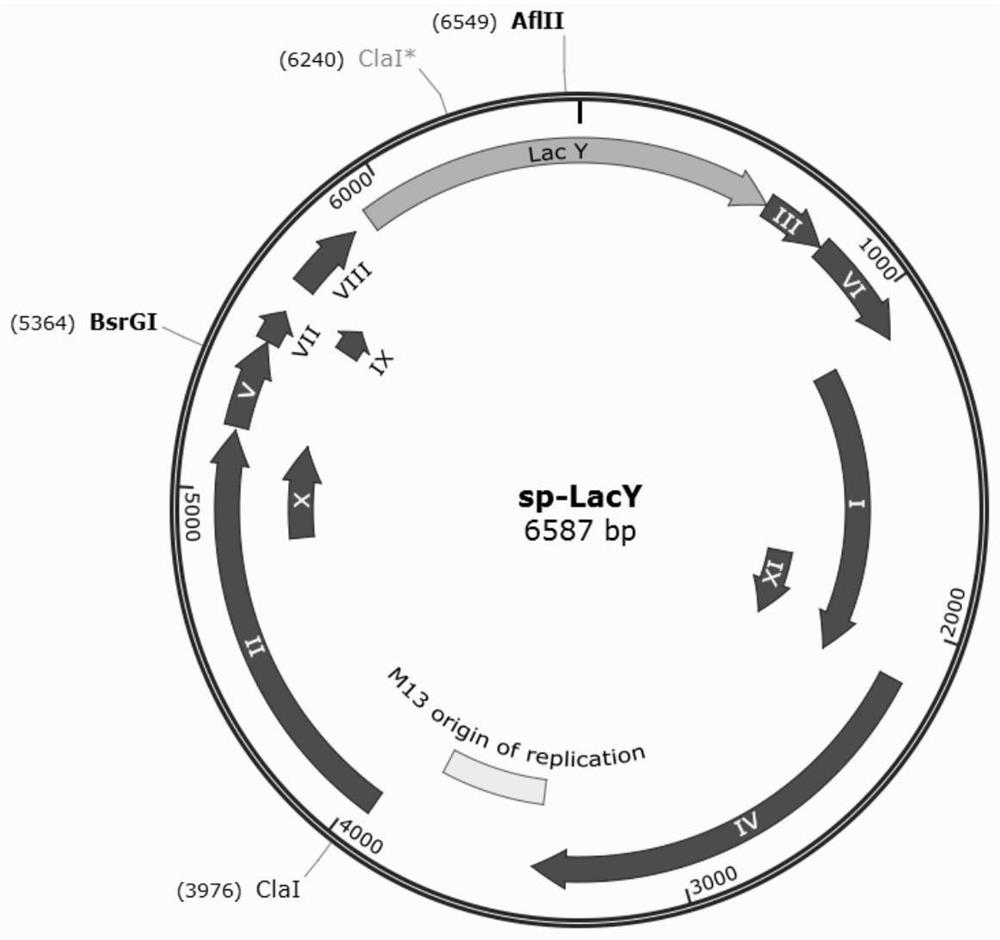
[0033] This embodiment provides a phage-assisted continuous directed evolution system of cello-oligosaccharide transporter, including: AV1 plasmid, AV2 plasmid and phage, the nucleic acid sequences of AV1 plasmid and AV2 plasmid are respectively SEQ ID NO.1 and SEQ ID NO.2 shown. Phage, that is, the phage SP-LacY in which the M13 phage gene gIII is replaced by the target gene LacY: PCR amplification of the M13 phage macroframe SP (without the gIII gene), PCR amplification of the wild-type lacY gene, and then using gibson to connect the M13 phage macroframe The SP and lacY genes form the SP-lacY double-stranded plasmid, and the gene map is as follows figure 1 As shown, the nucleic acid sequence is shown in SEQ ID NO.3.
[0034] The method for obtaining wild-type SP-LacY is as follows:
[0035]1) AV1 and AV2 co-transformed FM15 competent cells to obtain the host FM15-AV1 / AV2, which expresses on AV1 before cellobiose is transported into the cell without LacY protein (amino acid...
Embodiment 2
[0043] This example investigates the substrate specificity of LacY using the evolutionary system of Example 1, and adopts cellobiose, and the specific operations are as follows:
[0044] The initial phage was wild-type SP-lacY, titer 1×10 9 pfu / mL, the host is FM15-AV1 / AV2 (OD600=0.4), 1-2 uL of host bacteria are spotted in the center of LB soft agar (containing 1 mM cellobiose), and three spots are placed about 1 cm away from the host site. Phage droplets (1-2uL), three large phage fan-shaped spots can be seen the next day; the second round is similar, but the cellobiose concentration is reduced to 200uM, and the phage is diluted 100 times with the phage after the first round of evolution. board; for the following rounds, the third round is 40uM, the fourth round is 10uM, and the fifth round is 2uM. Until the formation of fan-shaped spots is difficult or new mutations no longer occur. Each round of sampling and sequencing was used to detect the accumulation of LacY mutation...
Embodiment 3
[0047] Evolution product LacY A177V Validation of transport activity using cellobiose:
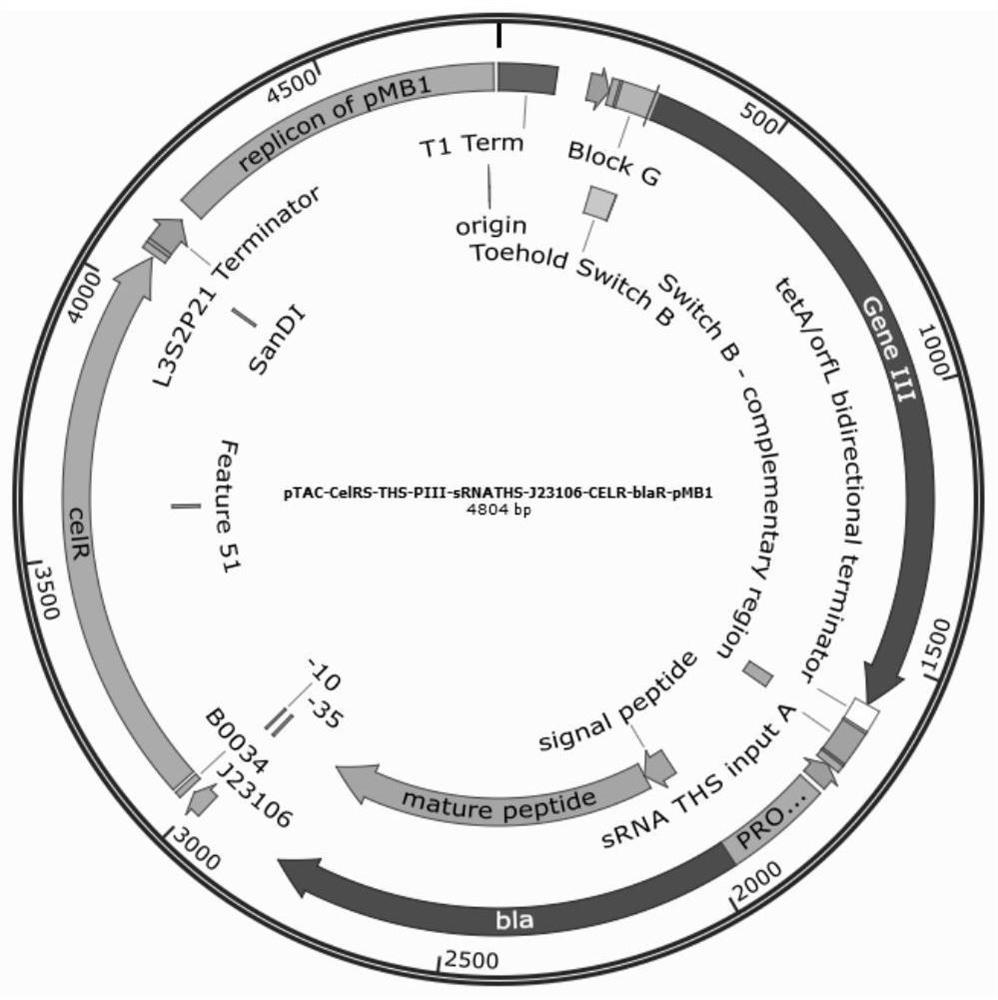
[0048] Construction of plasmid AV3-LacY wild and AV3-LacY A177V , which carry wild-type LacY or mutant LacY, respectively A177V , and both are constitutively expressed by promoter J23109 (AV3-LacY wild The plasmid map of Figure 7 shown, AV3-LacY wild Sequence structure such as SEQ ID NO.7); construct plasmid AV4 (plasmid map such as Figure 8 shown, SEQ ID NO. 8), which carries the reporter gene GFP, is initiated by pTAC-CelRS, and is controlled bi-levelly by CelR and the hairpin.
[0049] Plasmid AV3-LacY wild and AV3-LacY A177V Strain S1030-AV4 was transformed respectively to obtain engineering strain S1030-AV4 / AV3-LacY wild and S1030-AV4 / AV3-LacY A177V .
[0050] S1030-AV4 / AV3-LacY wild and S1030-AV4 / AV3-LacY A177V The cells were cultured in LB medium to log phase, 10uM cellobiose was added to induce GFP expression for 3h, centrifuged and washed twice with PBSbuffer, and t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com