Pseudomonas aeruginosa as well as construction method and application thereof
A technology of Pseudomonas aeruginosa and strains, applied in the field of genes, can solve problems such as no production correlation, and achieve the effect of increasing the production of rhamnolipids, improving the utilization rate of glycerol, and increasing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
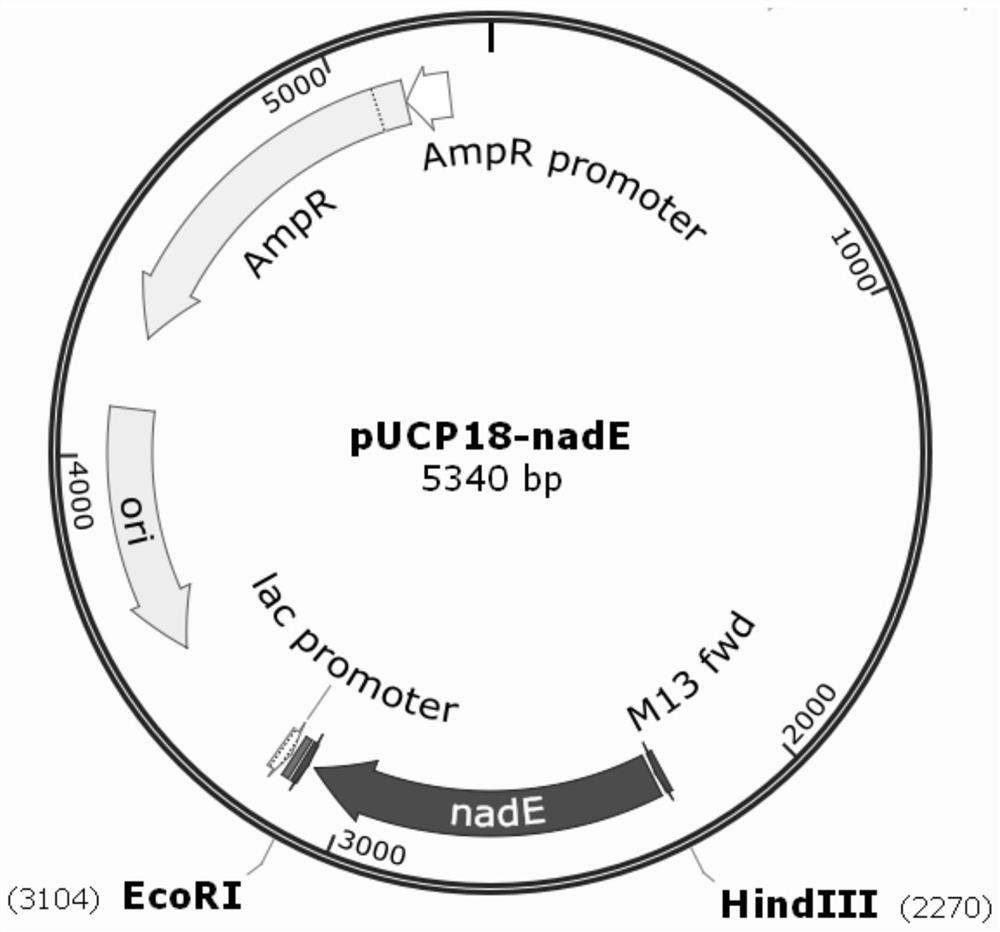
[0045] Embodiment 1: Construction of pUCP18-nadE vector
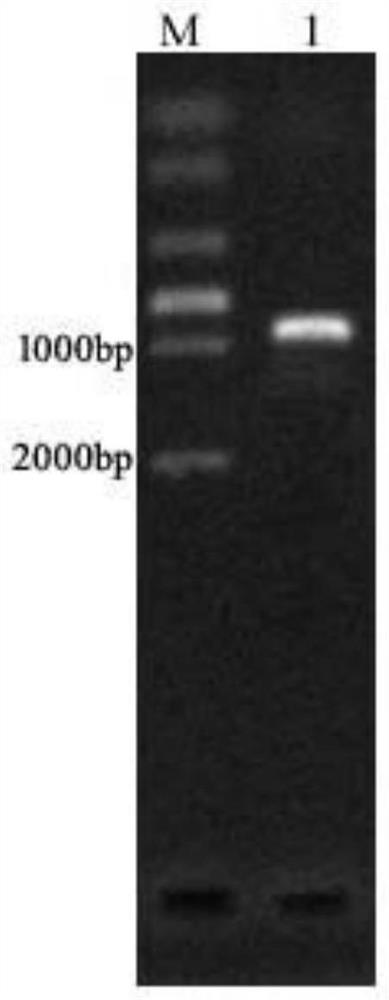
[0046]1. Use the genomic DNA of Pseudomonas aeruginosa KT1115 as a template, and use nadE-F and nadE-R as primers to perform PCR amplification to obtain a fragment containing the nadE gene. The nadE gene is shown in SEQ ID NO:3. The amino acid sequence of NAD+ synthetase encoded by nadE gene is shown in SEQ ID NO:4.
[0047] nadE-F: 5'-acgacggccagtgcc aagctt ATGCAACAGATCCAACGCG-3' (SEQ ID NO: 1)
[0048] The underlined sequence in SEQ ID NO:1 is the HindⅢ restriction enzyme recognition site;
[0049] nadE-R: 5'-tatgaccatgattac gaattc TCAGGGCGCCTTCGGCAG-3' (SEQ ID NO: 2)
[0050] The underlined sequence in SEQ ID NO: 2 is the recognition site for EcoRI digestion.
[0051] SEQ ID NO: 3:
[0052] ATGCAACAGATCCAACGCGACATCGCCCAAGCCCTGCAGGTCCAGCCGCCGTTCCAGTCGGAGGCCGACGTGCAGGCGCAGATCGCCCGGCGCATCGCCTTCATCCAGCAGTGCCTGAAGGATTCCGGGCTGAAGACCCTGGTGCTGGGGATCAGCGGCGGCGTCGACTCGCTCACCGCCGGCCTGCTGGCCCAGCGCGCCGTCGAGCAACTGCGCGAGCAG...
Embodiment 2
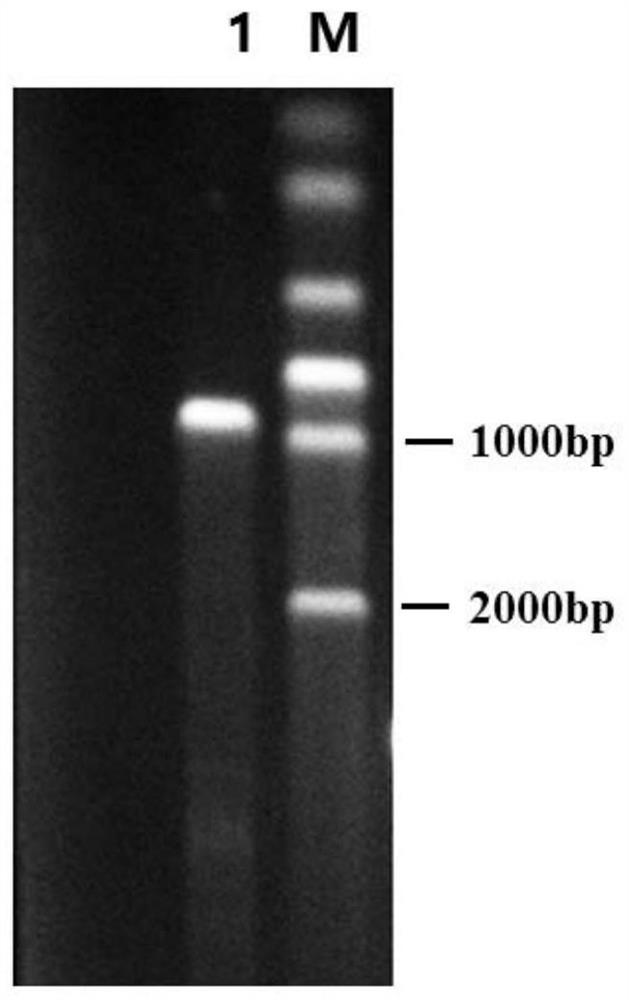
[0063] Embodiment 2: Construction of nadE gene overexpression strain KT1115-nadE
[0064] 1. Preparation of Pseudomonas aeruginosa KT1115 competent cells
[0065] (1) Pseudomonas aeruginosa KT1115 was inoculated in 2 mL of LB medium, and cultivated overnight at 30° C. and 200 rpm;
[0066] (2) According to the inoculum size of 1%, transfer the bacterial solution obtained in step (1) to 50mL LB medium, and cultivate it at 30°C and 200rpm until the OD600 is about 0.6;
[0067] (3) Centrifuge the bacterial solution obtained in step (2) at 8000 rpm and 4°C for 10 min, and take the supernatant;
[0068] (4) Resuspend the supernatant with 30mL pre-cooled 300mM sucrose solution, and wash 2-3 times;
[0069] (5) Finally, 0.5 mL of 300 mM sucrose solution was added to redissolve to complete the preparation of Pseudomonas aeruginosa KT1115 competent cells.
[0070] 2. Electrotransformation of Pseudomonas aeruginosa KT1115
[0071] (1) Take about 500ng of pUCP18-nadE vector and 100μL...
Embodiment 3
[0076] Embodiment 3: KT1115-nadE fermentation produces rhamnolipid
[0077] Fermentation medium composition: glycerin 40-100g / L, sodium nitrate 7.5g / L, peptone 5.0g / L, dipotassium hydrogen phosphate 2.5g / L, disodium hydrogen phosphate 2.5g / L, magnesium sulfate 0.5g / L, Calcium chloride 0.5g / L, the balance is water.
[0078] Pick a single colony of the recombinant bacterium KT1115-nadE obtained in Example 2 and inoculate it into 5 mL of LB medium at 30 ° C and 200 rpm for 12 hours to obtain a seed solution, and then inoculate 50 mL of the base with different contents according to the inoculum size of 6%. Carbon source in the fermentation medium, OD 600 0.3, 30°C, 200rpm shaking culture for 2 days, add IPTG with a final concentration of 1mM to induce the expression of NAD+ synthase, continue to ferment until the 7th day, end the fermentation, centrifuge at 8000rpm at room temperature for 10min, take the supernatant, and use the anthrone method to determine the supernatant The r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com