Recombinant human fucosyltransferase variant and application thereof
A technology of base transfer and mutation, applied in the field of protein mutants, can solve problems such as unsatisfactory clinical treatment, reduced enzyme activity, and poor enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1. Preparation of recombinant fucosyltransferase
[0030] 1.1 Plasmid preparation
[0031] Entrust Suzhou Junji Biotechnology Co., Ltd. to synthesize plasmid DNA and clone it into the vector pRM293 (pRM293 was obtained by transformation on the basis of plasmid pTT5, see Shi C. Purification and characterization of a recombinant G-protein-coupled receptor, Saccharomyces cerevisiae Ste2p, transiently expressed in HEK293 EBNA1 cells. Biochemistry. 2005;44(48):15705–15714.), respectively used to express FUT6, FUT10, FUT11 recombinant proteins (amino acid sequences were found in GenBank: M98825.1 or CCDS12152.1, or The SEQ ID NO.1 of this application; GenBank: AJ431184.1 or CCDS6088.1 and GenBank: BC036037.1 or CCDS7333.1, were constructed to express plasmids FUT6-pRM293, FUT10-pRM293, FUT11-pRM293. For specific operation methods, refer to " "Molecular Cloning Experiment Guide", the plasmid was first transformed into DH10B, sequenced, bacteria preserved and cultured ...
Embodiment 2
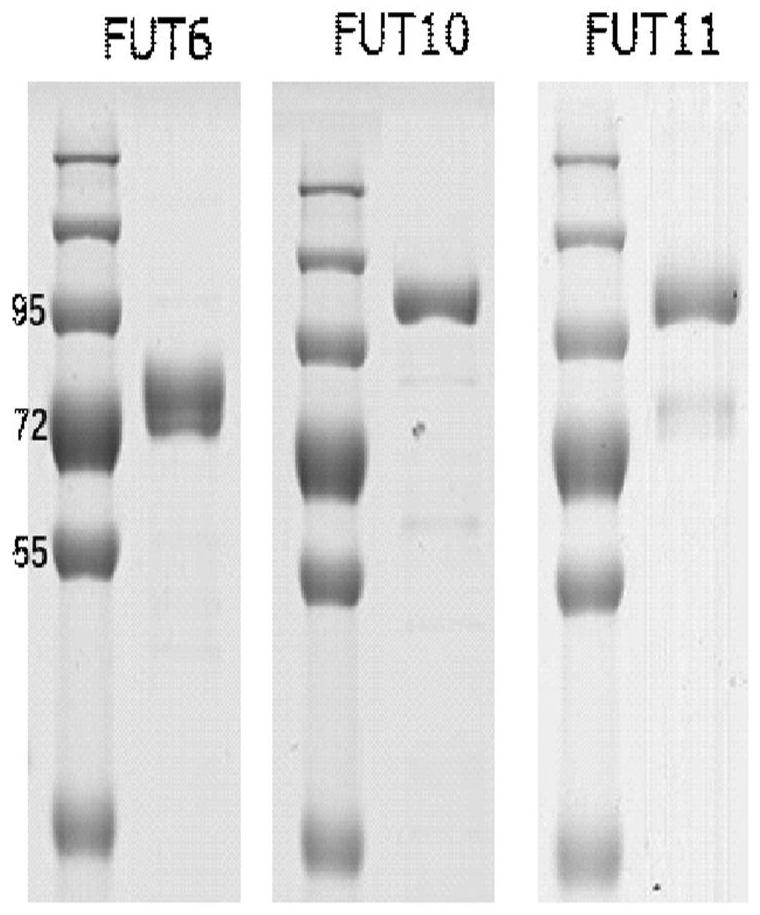
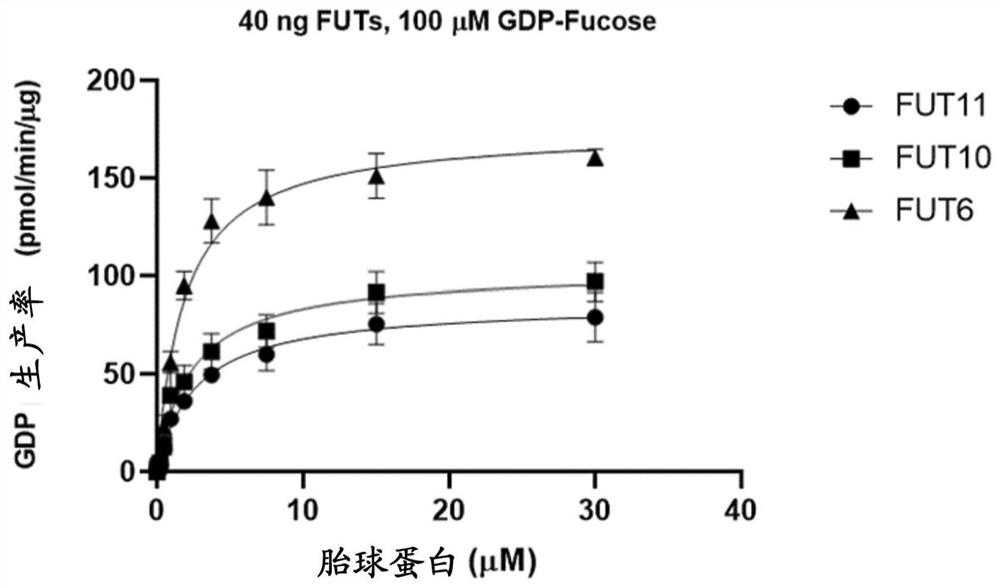
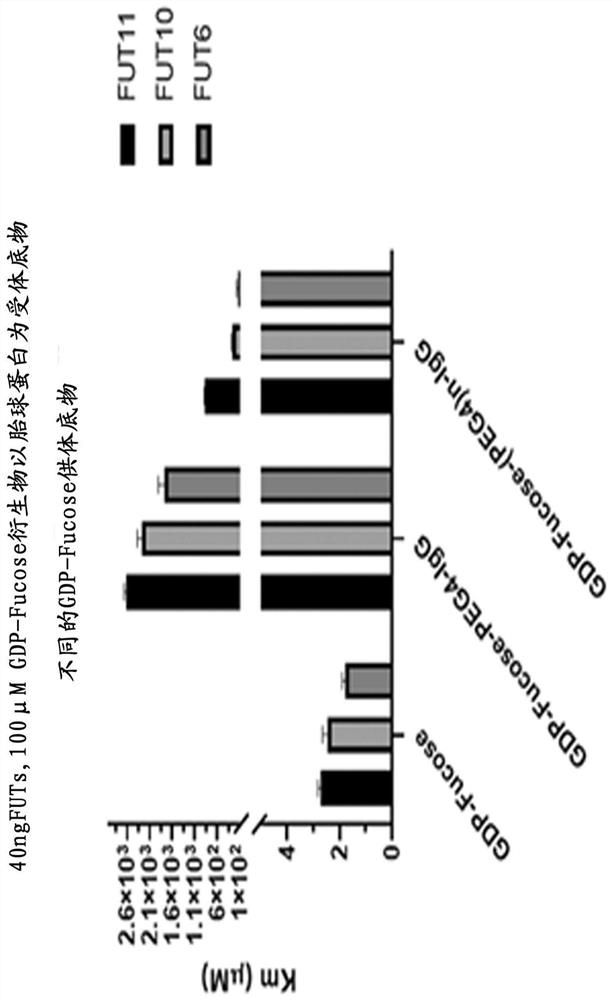
[0053] Example 2. Activity detection of recombinant fucosyltransferase
[0054] Purified FUT6, FUT10, FUT11 recombinases were analyzed and measured for enzymatic activity. The reaction buffer contained 40ng of recombinases and 100 µM ultrapure GDP-fucose (GDP-Fucose) (Promega Cat.#VA1097) as a donor substrate, and 40 µM fetuin (Promega Cat#V4961) as the acceptor substrate. All enzymatic reactions were performed in white 96-well plates in 25 µL volumes. Incubate for 60 minutes at room temperature. The GDP glycosyltransferase assay was performed as described in the manual (Promega GDP-Glo™ Glycosyltransferase Assay).
[0055] Contains 40 ng of recombinant fucosyltransferase and 100 µM GDP-fucose as donor substrate and double dilution (40 µM) of fetuin as acceptor substrate in a 25 µl reaction volume of PBS buffer and incubate at 37°C for 30 minutes. Then add 25 µl GDP Detection Reagent at room temperature, incubate for 60 minutes, put into GloMax® 96 microplate luminescence ...
Embodiment 3
[0062] Example 3. Preparation and activity detection of mutant FUT6 recombinase
[0063] There are currently no studies showing how fucosyltransferases recognize donor substrates, nor data for structural elucidation. According to the above experiments, it is confirmed that FUT6 can recognize not only small molecule donor substrates, but also macromolecular donor substrates. To improve the ability of FUT6 to recognize macromolecular donor substrates, the amino acid sequence analysis of FUT6 was performed. There are 11 human-derived fucosyltransferases, which belong to the Glycosyltransferse family 10 (Glycosyltransferse family 10) with the prokaryotic fucosyltransferases, but there is no obvious homology in the amino acid primary sequence. Sequence comparison between FUT6 and the alfa-1,3 fucosyltransferase of Helicobacter pylori revealed that FUT6 has 7 "gap" (Gap), that is, in the wild-type amino acid sequence shown in SEQID NO. 1 Amino acid 109 to amino acid 110, amino aci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com