Flory polyglycoside anabolism combined gene and combined enzyme
A technology of synthesizing metabolism and combining enzymes, which is applied in the field of genetic engineering and can solve problems such as less research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
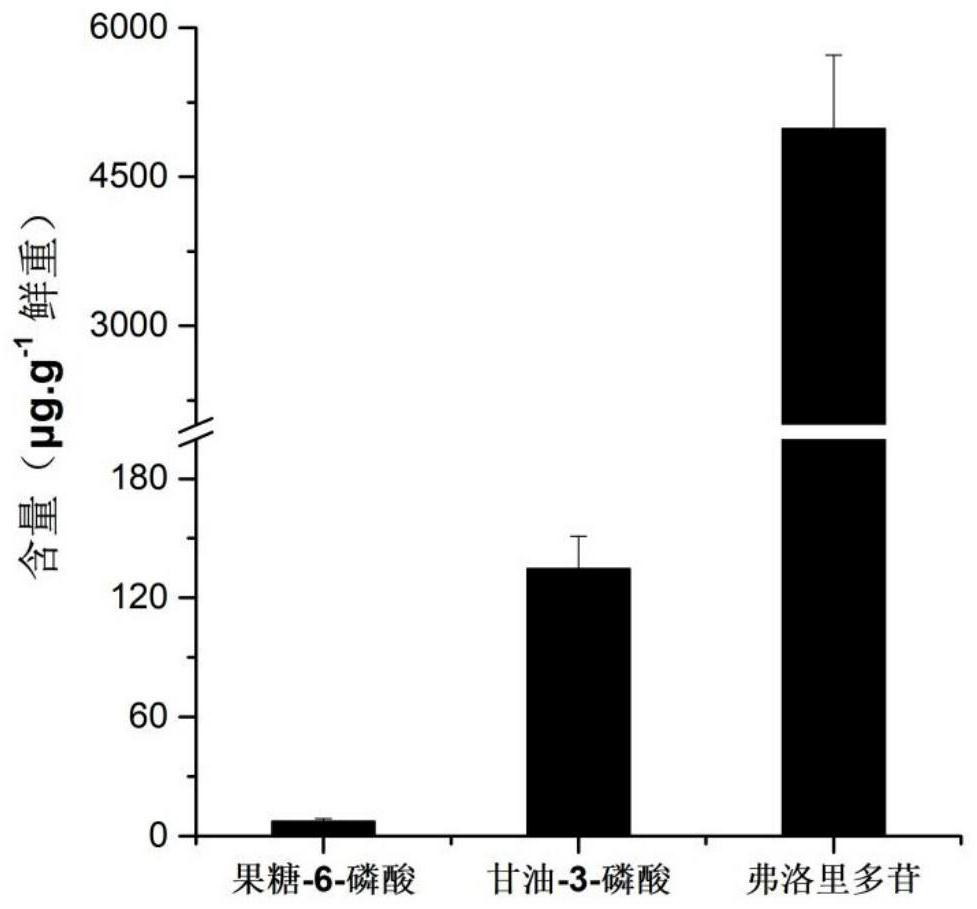
[0035] The sample is the fronds of wild altar Porphyra collected in Dongjia Island, Fujian Province on December 11, 2019. The Porphyra altar was transferred to an indoor aerated culture system, and the PES medium (Provasoli, L., 1968) was used to recover the culture at 21°C for 48 hours in the dark. After resuming the culture, put Porphyra altar at 21°C, 50mol·m -2 the s -1 Under the condition of light (12h light, 12h dark), culture medium was changed every 2 days. On the 5th day of cultivation, when the light was 6 hours, Porphyra altar with almost the same size was selected for gene analysis and content determination.
Embodiment 2
[0036] Example 2 Research on Floridoside Synthesis and Metabolism Pathway
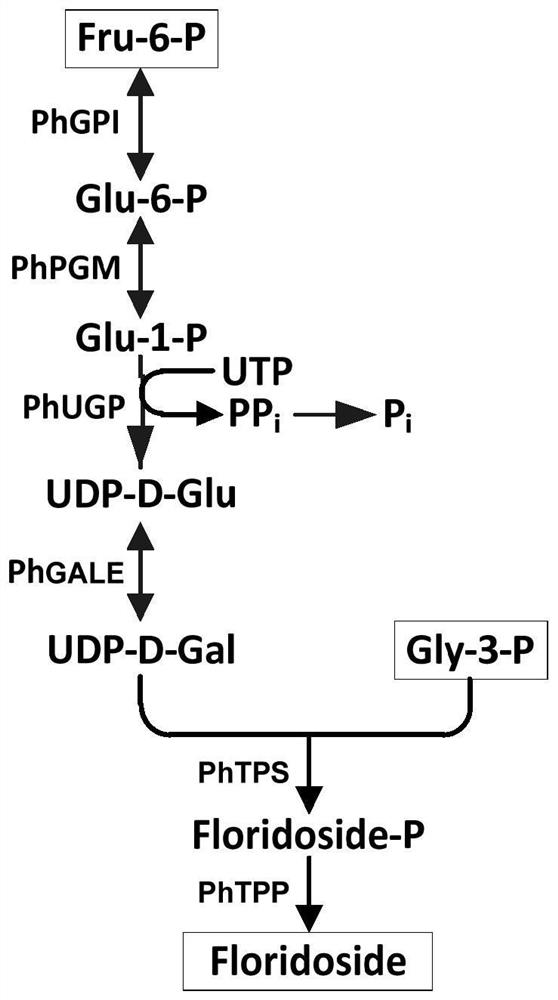
[0037] Synthetic and metabolic pathways of floridoside in laver figure 1 As shown, fructose-6-phosphate (Fru-6-P) is converted by phosphoglucose isomerase (PhPGI) to glucose-6-phosphate (Glu-6-P), which is then converted by glucose phosphomutase (PhPGM) Glucose-1-phosphate (Glu-1-P). Under the catalysis of UDPG pyrophosphorylase (PhUGP), Glu-1-P reacts with UTP to generate UDP-glucose (UDP-Glu). UDP-glucose is isomerized to UDP-galactose (UDP-Gal) under the catalysis of UDP-glucose 4-isomerase (PhGALE). Then UDP-galactose and glycerol-3-phosphate (Gly-3-P) synthesize floridoside under the catalysis of trehalose phosphate synthase (PhTPS) and trehalose phosphate phosphorylase (PhTPP).
Embodiment 3
[0038] Example 3 Gene Analysis
[0039] (1) From NCBI (https: / / www.ncbi.nlm.nih.gov / ), MGU
[0040] (https: / / marinegenomics.oist.jp / algae / gallery) and Ensembl
[0041] (http: / / plants.ensembl.org / index.html) Download the homologous sequences involved in the floridoside metabolic pathway from the database, and save them in fasta format;
[0042] (2) Utilize Secure FX and Putty software to connect to the Porphyra genome database on the main server, and run the command "formatdb-iOneKP.faspF" in Putty to format the database;
[0043] (3) Use Secure FX to upload the protein sequence file downloaded to the local computer to the formatted database folder;
[0044] (4) Use the "blastall-p tblastn-d(sequence name)-i(searchsequence)-o out-(gene name)-FF-e 1e-5" command in Putty to retrieve the sequence with amino acid as query, The result file is opened in .txt format and saved to the local computer;
[0045] (5) Use Editplus (https: / / www.editplus.com / ), perl and Tbtools software to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com