Method for preparing inositol by multi-enzyme reaction system expressed by edible microorganisms
A multi-enzyme system and expression method technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve problems such as complex purification processes, and achieve the effect of reducing production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
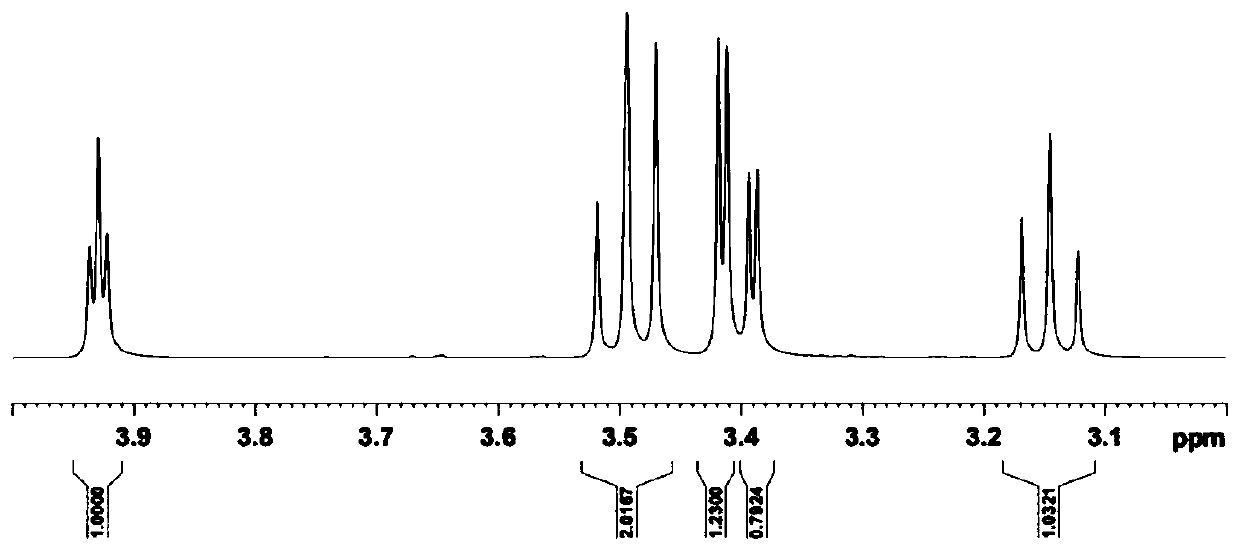
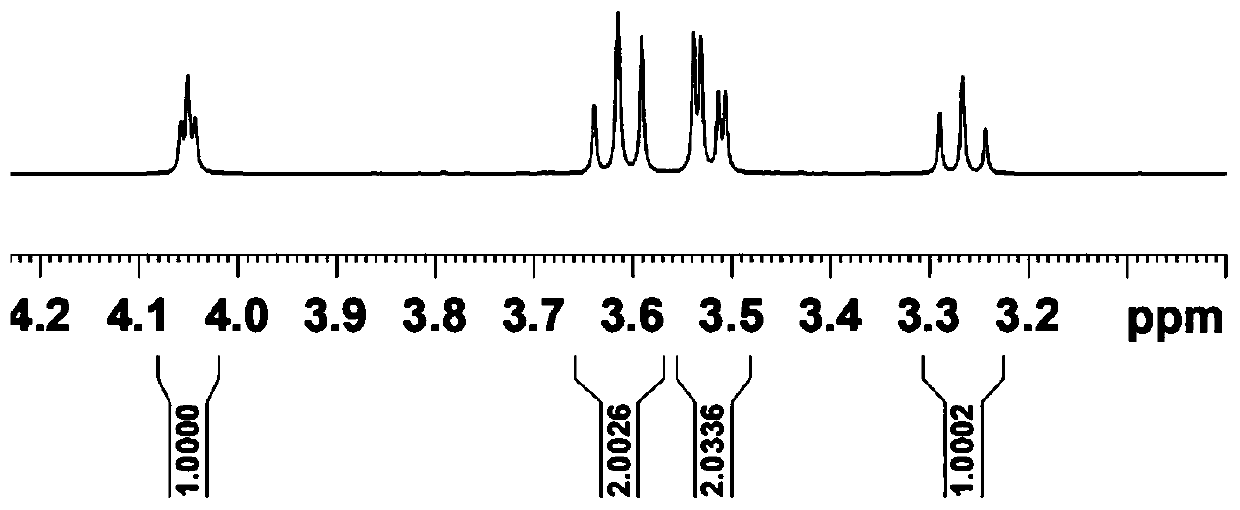
Image
Examples
Embodiment 1
[0034] Embodiment 1: Lactic acid bacteria prepare multienzyme
[0035] The five genes whose KEGG numbers are ST0928, TM1168, TM0769, AF1794 and TM1415 were obtained from the official website of ATCC (www.atcc.org). Different primers were used to obtain from the corresponding genomic DNA by PCR, through Simple Cloning (You,C.,et al.(2012). "Simple Cloning via DirectTransformation of PCR Product (DNA Multimer) to Escherichia coli and Bacillus subtilis." Appl.Environ.Microbiol.78(5):1593-1595.) cloned into the pNZ9530 vector to obtain the corresponding gene expression vector, and then respectively transformed into lactic acid bacteria NZ9000 for protein expression. After detection, the target enzyme live negative.
Embodiment 2
[0036] Embodiment 2: Saccharomyces cerevisiae prepares multienzyme
[0037] The five genes were cloned into the pYES2 vector by the same method as in Example 1 to obtain the corresponding gene expression vectors, and then transformed into Saccharomyces cerevisiae Y187 for protein expression. The target enzyme activity was negative after detection.
Embodiment 3
[0038] Embodiment 3: Pichia pastoris prepares multienzyme
[0039] Using the same method as in Example 1, the five genes were cloned into the pPIC9K vector to obtain the corresponding gene expression vectors, and then transformed into Pichia pastoris GS115 for protein expression. The target enzyme activity was tested negative.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com