Aminopeptidase Amp0279 sourced from lysinibacillus sphaericus C3-41 as well as recombinant strain and application of aminopeptidase Amp0279
A technology of pht43-amp0279 and Bacillus lysinus, which is applied in the field of protein engineering and genetic engineering, can solve the problem of low yield of other proteins and achieve the effect of stable enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1: Aminopeptidase AMP0279 Structure and Function Prediction and Gene Sequence
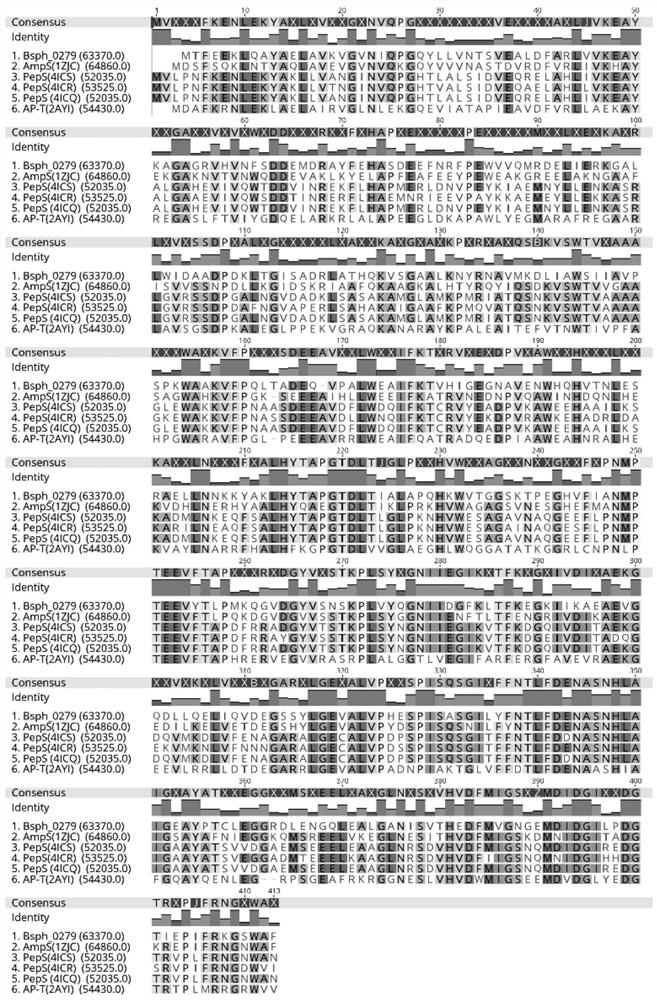
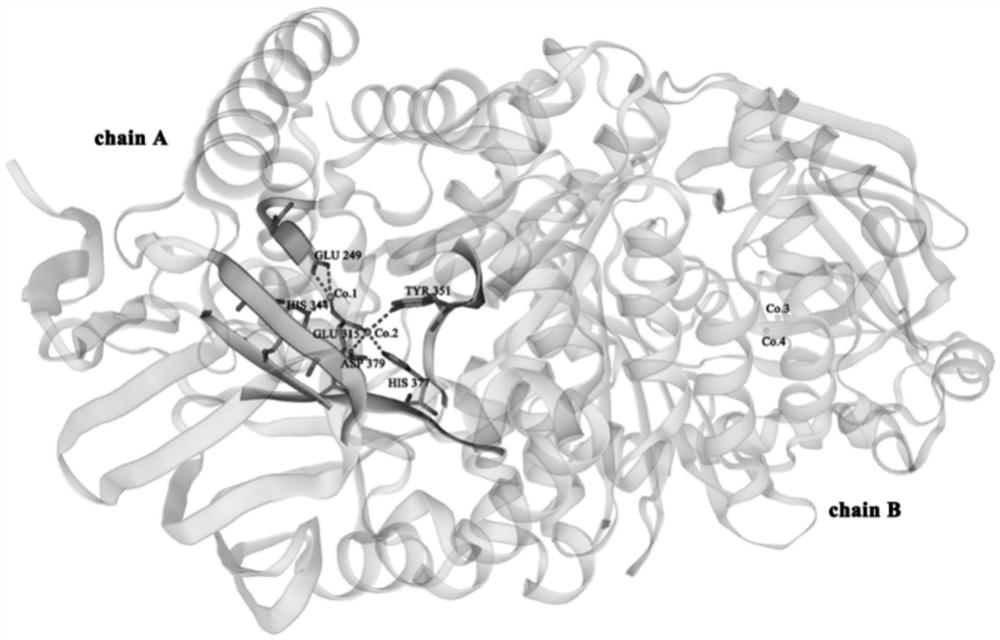
[0065] The protein AMP0279 encoded by the AMP0279 gene (300199..301428) may be predicted according to the spherical lysine Bacillus C3-41 genome sequence (CP000817), which may predict the protein AMP0279 encoded by the AMP0279 gene (300199..301428). The sequence alignment has the highest similarity (51%) (51%) (51%) (51%) (51%) (51%) (51%) (51%). figure 1 . 3D modeling analysis found that it may be a dimer structure with four cobalt ion binding sites, which may belong to the M29 family (see figure 2 . figure 2 Is the AMP0279 3D Structure Model (SWSS-Model). AMP0279 is a homologous dimer. The chart is chain a chain A, leaning on the right is chain B, 4CO 2+ For its active center, labeled a polka dot. The six active sites (Glu 249, Glu 315, His 344, TYR 351, HIS 377, and ASP 379) are respectively coupled to two COs around the chain A. 2+ . The primer with enzyme dug bit BamHI and SACI is d...
Embodiment 2
[0066] Example 2: Recombinant expression plasmid PET28A-AMP0279 and recombinant BL21 (PET28A-AMP0279)
[0067] The amino peptidase AMP0279 gene 5 'end and 3' ends are introduced into restriction endonuclease BamHI and SACI, with restriction endonuclease BamHi and SACI to complete the aminopeptidase AMP0279 gene and E. coli induced expression vector PET28A. Digestion, then use the ligase to connect both, construct the recombinant expression plasmid PET28A-AMP0279. The recombinant expression plasmid was transformed into E. coli BL21, screened by PCR verification, and the positive clonal strain BL21 (PET28A-AMP0279) was screened, and plasmid sequence verification was extracted.
Embodiment 3
[0068] Example 3: Nickel pillar and chromatography method Purified aminogenase amp0279
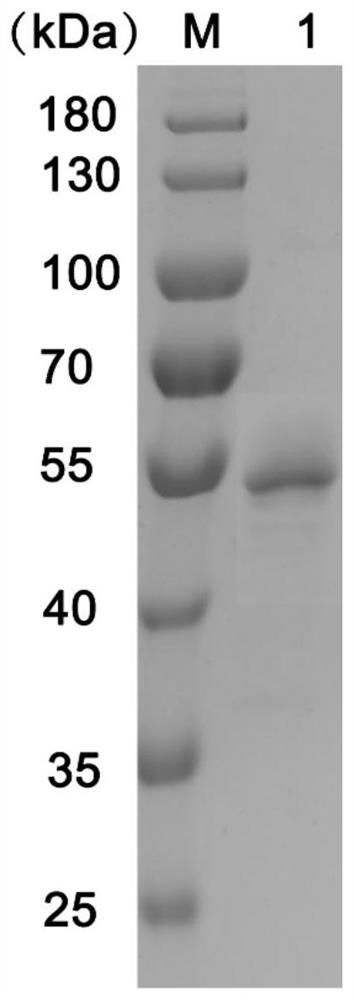
[0069] The recombinant bacteria were seeded in 5 mL lb (50 μg / ml kanamycin), 37 ° C, 220 rpm overnight culture obtained seed fluid. The seed fluid is added to the proportion of 1: 100 (50 μg / ml kanamycin), and the culture is cultured to the OD. 600 When = 0.6-0.8, IPTG (1 mm at a final concentration) was added to low temperature and slow-speed induction (25 ° C, 160 rpm) for 5 h. Centrifugation (5000g, 10min) is bacterial, abandoned. Ultrasonic crushes, 4 ° C, 12000 g, 30 min after centrifugation, reserved supernatant for protein nickel-column affinity chromatography. After dialysis, the protein concentration was detected with a BCA kit. The known concentration of pure ammoniapinamepase AMP0279 can be detected by SDS-PAGE to a single strip of about 55 kD (purified after the detection of AMP0279) image 3 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com