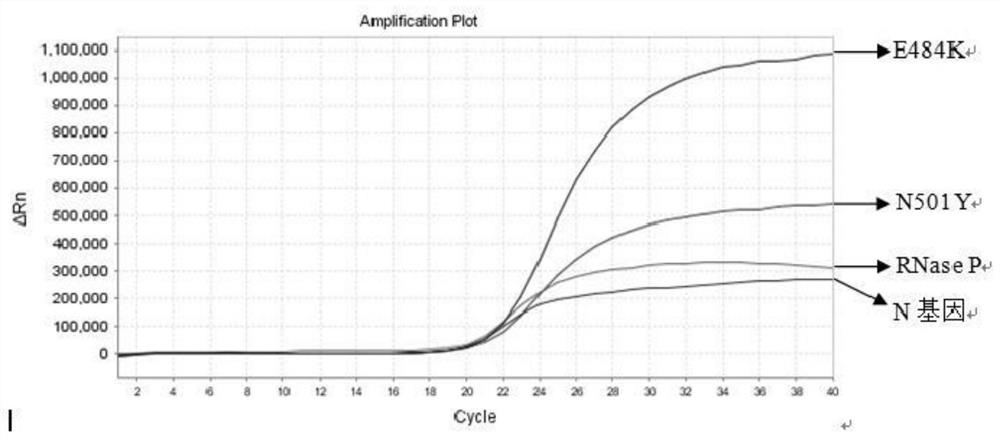
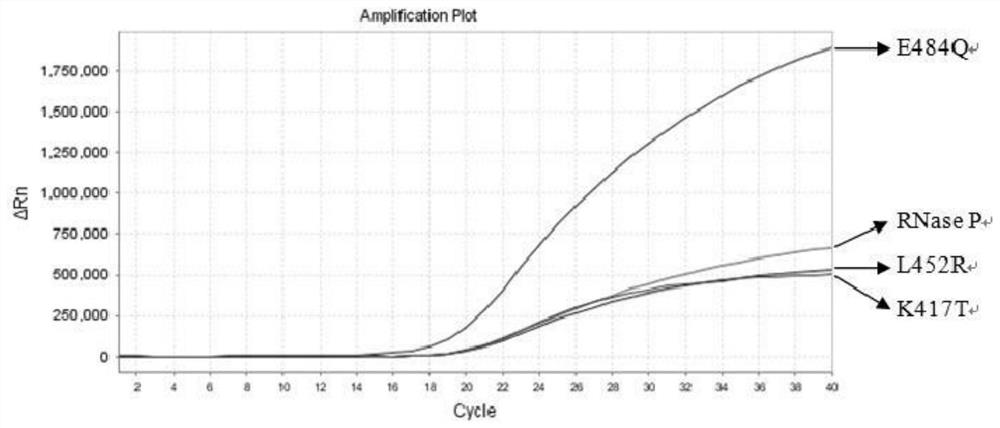
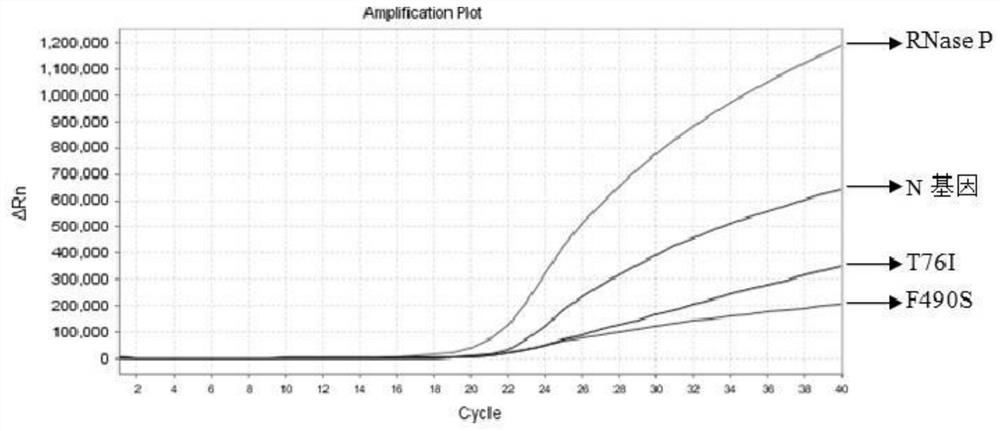
Nucleic acid composition, kit and method for simultaneously detecting multiple mutant strains of SARS-CoV-2
A nucleic acid composition and coronavirus technology, which is applied in the field of nucleic acid detection, can solve the problems of inconvenient virus monitoring and disease treatment, and the inability to quickly detect new coronaviruses, and achieve stable and reliable detection results, stable and reliable experimental results, and cost-saving detection. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] A nucleic acid composition for simultaneously detecting multiple mutant strains of novel coronavirus
[0068] Synthetic primers and probes were designed for the nucleic acid sequences of the eight mutants:
[0069] According to the nucleic acid sequences of the new coronavirus (SARS-CoV-2) Alpha, Beta, Gamma, Eta, Delta, lota, Kappa, Lambda mutant strains published by NCBI, eleven sets of specific primers were designed using Oligo7.0 primer design software and probe. The sequences of primers and probes are shown in Table 1.
[0070] Table 1 Eleven sets of primer probe sequences
[0071]
[0072]
[0073]
[0074]
[0075] The 5' end of each probe sequence is modified with a fluorescent reporter group, and the 3' end of each probe sequence is modified with a fluorescent quencher group. Wherein, the fluorescent reporting group is selected from one of FAM, JOE, VIC, HEX, ROX, TEXAS RED and CY5, and the fluorescent quenching group is selected from one of BHQ1...
Embodiment 2
[0081] A kit for simultaneously detecting multiple mutant strains of novel coronavirus
[0082] One or more of the novel coronavirus (SARS-CoV-2) Alpha, Beta, Gamma, Eta, Delta, lota, Kappa, and Lambda mutant strains published by NCBI can be detected simultaneously.
[0083] The kit includes the following reagents: primers and probes for one or several sites in the nucleic acid composition obtained in Example 1; enzyme mixture; PCR reaction reagents.
[0084] The enzyme mixture includes DNA polymerase, reverse transcriptase, and Rnasin. The DNA polymerase is a hot-start Taq enzyme, and the reverse transcriptase includes c-MMLV reverse transcriptase or AMV reverse transcriptase. In this example, c-MMLV reverse transcriptase is used.
[0085] The PCR reaction reagents include PCR buffer, dNTPs and cations, the PCR buffer is 10×PCR Buffer, and the cations are derived from MgCl 2 .
[0086] Preparation of reagents: take reaction solutions Ⅰ, Ⅱ, Ⅲ, Ⅳ: N×19 μL; enzyme mixture: N×1...
Embodiment 3
[0102] A method for simultaneously detecting multiple mutant strains of novel coronavirus, comprising the following steps:
[0103] S1. Collect and extract the RNA of the test sample:
[0104] For sample collection, refer to the throat swab or sputum collection method in the "Technical Guidelines for Laboratory Testing of Novel Coronavirus Pneumonia".
[0105] Pharyngeal swab sample: Pass the swab over the base of the tongue, wipe the pharyngeal tonsils on both sides of the subject with a little force back and forth, and then wipe up and down the back wall of the pharynx, dip the swab head into the tube containing the virus preservation solution, and discard the tail. Tighten the tube cap.
[0106] Sputum sample: After the patient coughs deeply, collect the coughed up sputum into a tube containing virus preservation solution, and tighten the cap of the tube tightly.
[0107] In this embodiment, the throat swab collection method is adopted.
[0108] A nucleic acid extraction...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com