Method and screening model for screening small-molecule inhibitor of main protease of new coronavirus
A small molecule inhibitor and main protease technology, applied in the field of medicine and biology, can solve the problems of long screening period, limited application, cumbersome operation, etc., and achieve the effect of sensitive detection, easy operation and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1. Prokaryotic expression and isolation and purification of the new coronavirus Mpro recombinant protein
[0038] Using Escherichia coli prokaryotic expression technology, the codon-optimized Mpro gene ( figure 1) was connected to the pET-21a (+) expression vector to construct the recombinant plasmid Mpro-pET-21a. Then the recombinant plasmid was transformed into E. coli Rosetta (DE3) competent cells, and the recombinants were screened by ampicillin resistance. The recombinants were inoculated into 1 liter of LB liquid medium (containing 100 μg / mL ampicillin), cultured at 37° C. for 7 hours, added with 0.2 mM IPTG, and induced at 30° C. for 8 hours. After the cells are disrupted by ultrasonic method, the lysed supernatant is separated and purified by HisTrap affinity chromatography column. The purified Mpro has an apparent molecular weight of 34kDa, and only one formylmethionine remains at the amino terminal, and polyhistidine is fused to the carboxyl terminal....
Embodiment 2
[0045] Example 2. Biological Activity Identification of New Coronavirus Mpro Recombinant Protein
[0046] 1. Dilute 10 μM MCA (7-methoxycoumarin, 7-methoxycoumarin-4-acetic acid, MCA) in TBS solution (50mM Tris, 150mM NaCl pH8.0) to 5 concentrations by 2 times For gradient, the wells containing only TBS solution were used as negative control wells. The above MCA dilution was added to a 384-well plate at 50 μL / well, and the total amount of MCA in each well was 0, 31.25, 62.5, 125, 250, and 500 pmol, and the automatic gain mode was set to detect the relative fluorescence intensity with a multi-functional microplate reader Value (relative fluorescence unit, RFU). According to the total amount of MCA in each well and the ΔRFU value (ΔRFU=RFU MCA -RFU 0 ) to fit the regression equation, draw the MCA fluorescence intensity standard curve ( figure 2 B).
[0047] 2. Add 2mM MCA-Substrate (MCA-AVLQSGFR-Lys(Dnp)-Lys-NH 2 ) was diluted to 10 μM with TBS solution, and Mpro was adde...
Embodiment 3
[0050] Example 3. The hydrolysis of the new coronavirus Mpro recombinant protein to the FITC-S-Biotin substrate
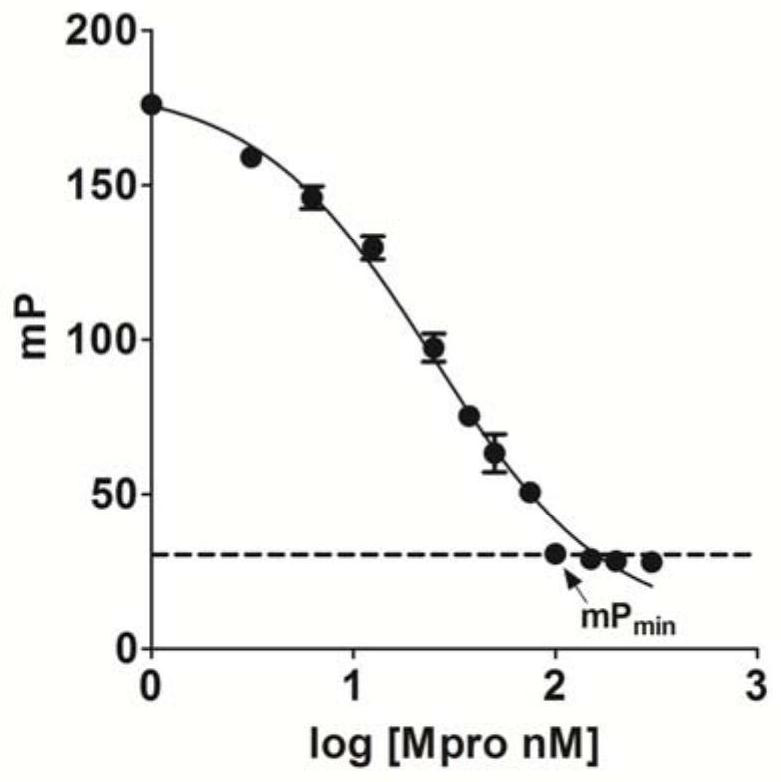
[0051] Dilute 2mM FITC-S-Biotin to 60nM with fluorescence polarization reaction solution (10mM Tris, 50mM NaCl, 1mM DTTpH8.0), add 20μL / well into a 384-well plate, and then add 0, 25, 50, 100, 150, 175, 200, 300, 400, 500, 600, 700nM Mpro, 30μL per well, set 3 sets of duplicate wells for each group, incubate at room temperature for 20min, then add 300nM avidin reaction solution at 10μL / well, incubate at room temperature in the dark After 5 min, the mP value was detected with a multifunctional microplate reader.
[0052] The Mpro hydrolysis reaction curve shows that 200nM Mpro can fully hydrolyze the FITC-S-Biotin substrate into the product FITC-AVLQ( image 3 ).
PUM
| Property | Measurement | Unit |
|---|---|---|
| Apparent molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com