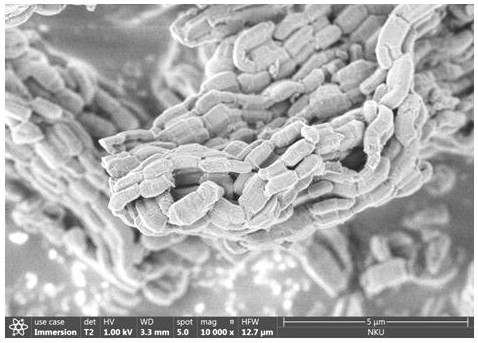
Preparation method of glycoprotein microreactor for boron affinity surface imprinting of mesoporous molecular sieve
A technology of mesoporous molecular sieve and microreactor, applied in the field of glycoproteomics analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The preparation of mesoporous molecular sieve boron affinity surface imprinted polymer (SBA-15@MIP), this example includes the following five steps:
[0035] (1) Pretreatment of SBA-15 molecular sieve
[0036] SBA-15 (Shanghai Aladdin Reagent Co., Ltd.) was activated in HCl solution to obtain more hydroxyl groups. The specific operation is as follows, disperse 200 mg of SBA-15 into 8 mL of 6 M HCl, and stir at room temperature for 10 hours. Then, the supernatant was discarded by centrifugation, and the SBA-15 was washed five times with deionized water until the pH of the supernatant was neutral. Secondly, the acidified SBA-15 was vacuum-dried at room temperature, and then vacuum-dried at 110° C. for 6 hours.
[0037] (2) Amino functionalization of SBA-15 mesoporous molecular sieve materials
[0038] Amino-functionalized SBA-15 was prepared using APTES (3-aminopropyltriethoxysilane). First, 200 mg of SBA-15 was uniformly dispersed in a 100 mL glass flask containing 5...
Embodiment 2
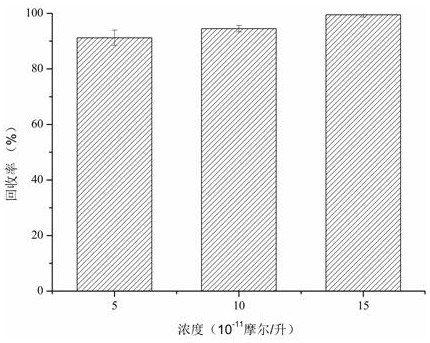
[0047] The extraction performance of SBA-15@MIP on template molecules in actual samples, such as figure 2 .
[0048] To further evaluate the extraction performance of the obtained mesoporous molecular sieve boron affinity surface-imprinted polymer (SBA-15@MIP) for template molecules in real samples, SBA-15@MIP was used to detect N-acetyl Extraction recovery of neuraminic acid. The present invention investigates the extraction performance of normal human serum samples added with different concentrations of N-acetylneuraminic acid. The specific operation steps are as follows:
[0049] (1) Synthesize SBA-15@MIP with the above method (Example 1).
[0050] (2) In this experiment, SBA-15@MIP was coated on the surface of glassy carbon electrode (GCE) by drop coating method. The specific steps are as follows: First, 1.0, 0.3 and 0.05 μM of GCE alumina slurry were applied on the polishing cloth respectively. The glassy carbon electrode was polished, and then ultrasonically cleaned...
Embodiment 3
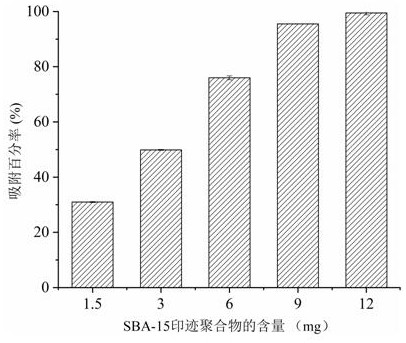
[0057] The effect of the amount of SBA-15@MIP added on protein adsorption in serum samples, such as image 3 . Specific steps are as follows:
[0058] (1) Weigh 1.5-12 mg of SBA-15@MIP and place it in a centrifuge tube. Afterwards, 2 μL of serum to be treated was added, and 48 μL of Tris-HCl solution (pH 8.0, 50 mM) was added to wet the material, and the supernatant was obtained by centrifugation after shaking (room temperature).
[0059] (2) Quantitative analysis was carried out according to the Bradford method. Mix the dye Coomassie blue G-250 and the sample to be tested at a volume ratio of 10:1, and then incubate at room temperature for 2 minutes to facilitate thorough mixing, and then measure the absorbance at 595 nm with a UV-3310 spectrophotometer, record . Finally, the percent protein adsorbed was calculated from the standard curve of the protein.
[0060] According to the obtained data, draw the corresponding image 3 . When 2 μL of human serum samples were add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com