Detection method for detecting bovine coronavirus
A coronavirus, a technology to be detected, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, and resistance to vector-borne diseases, etc., can solve the problem of qualitative and quantitative detection of untraceable mutations, complex processes, and low accuracy, etc. problems, to achieve high specificity, high sensitivity, and high precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The design and synthesis of embodiment 1, BCoVddPCR detection primer and probe
[0029] Referring to the whole genome sequence of BCoV recorded in GenBank in NCBI, the N gene of BCoV was selected as the target gene, and a pair of specific primers and probes were designed using Primer premier 5.0 software, which were synthesized by Shanghai Bioengineering Co., Ltd. The specific sequence is as follows:
[0030] Upstream primer: 5'-GATCTACTTCACGCGCATCC-3' (SEQ ID NO.2);
[0031] Downstream primer: 5'-GTGGCTTAGTGGCATCCTTG-3' (SEQ ID NO.3);
[0032] Probe: 5'-FAM-TGGCTCTACTGCGCGATCCTGCA-BHQ1-3' (SEQ ID NO.4);
Embodiment 2
[0033] The preparation of embodiment 2, BCoV standard positive plasmid
[0034] Total virus RNA was extracted by Trizol method, and reverse-transcribed into cDNA using HiFiScript cDNA Synthesis Kit reagent (Kangwei Century Biotechnology Co., Ltd.). Use 2× Hieff Canace Gold PCR Master Mix high-fidelity enzyme premix (Shanghai Yisheng Biological Co., Ltd.) to amplify the BCoV N gene and perform 1% agarose gel electrophoresis, use The Gel Extraction Kit agarose gel recovery kit (Shanghai Yisheng Biological Co., Ltd.) was used to recover the target gene. use Easy Vector System I Vector Kit (Promega) ligated the recovered product, and transformed the ligated product into Escherichia coli DH5α competent cells by heat shock. Use 2x Taq PCR Mix reagent (Tiangen Biochemical Technology (Beijing) Co., Ltd.) for bacterial liquid PCR identification, and use positive bacterial liquid Plasmid Mini Kit (Shanghai Yisheng Biological Co., Ltd.) was used to extract the plasmid, and the extr...
Embodiment 3
[0035] Embodiment 3, the establishment of BCoV TaqMan fluorescent quantitative PCR detection method
[0036] Using the primer set and probe described in Example 1, and using the standard positive plasmid prepared in Example 2 as a template, configure a qPCR 20 μL reaction system as follows:
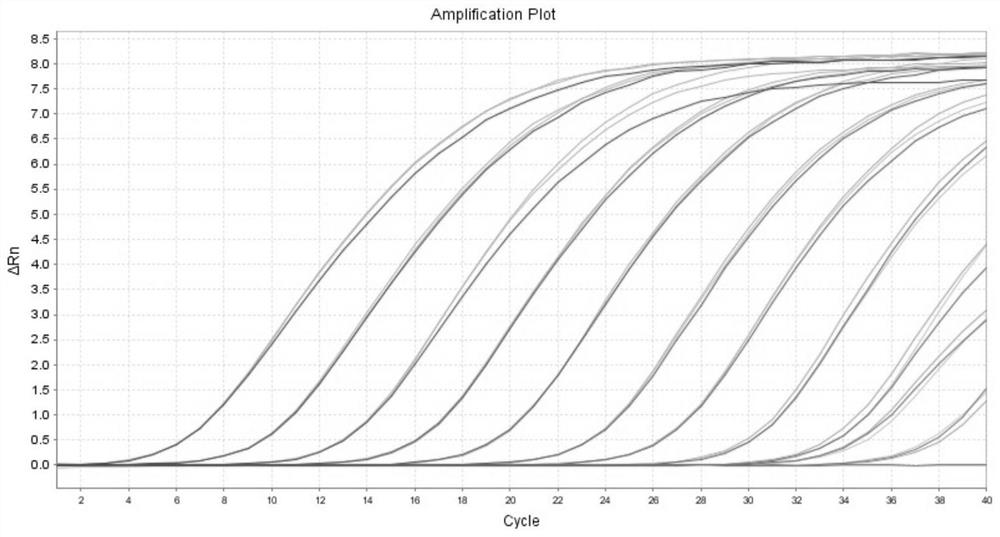
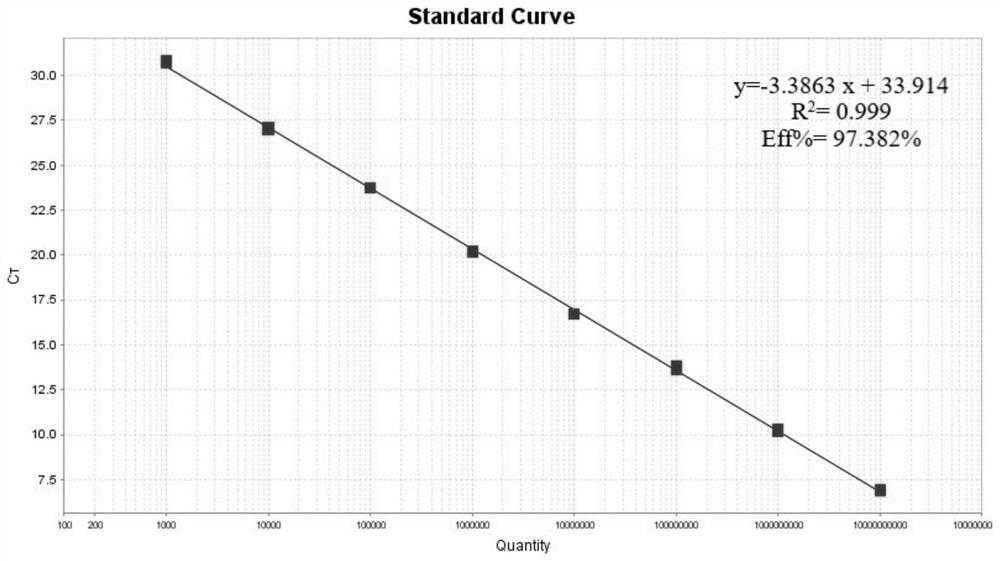
[0037] SuperRealPreMix (Probe) 10μL, upstream primer (10pmol / L) 0.8μL, downstream primer (10pmol / L) 0.8μL, probe (10pmol / L) 0.9μL, 50×ROX Reference Dye 0.2μL, RNase-free waste 5.3μL , template 2 μL; qPCR reaction conditions are: 95 ° C pre-denaturation 15 min; 95 ° C denaturation 10 s, 60 ° C annealing 30 s, a total of 40 cycles; 1.0 × 10 10 ~1.0×10 0 Copies / μL standard positive plasmid was used as a template for qPCR reaction, and a negative control was set up. The obtained amplification curve was as follows: figure 1 As shown, the template concentration from left to right is 1.0×10 10 ~1.0×10 0 copies / μL, the result shows that the minimum detection limit of this method is 10copies / μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com