The core sequence, primers and application of SNP markers related to high pH resistance of Chinese prawn
A Chinese shrimp, marker technology, applied in recombinant DNA technology, biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of lack of SNP markers, and achieve the effect of speeding up the breeding process and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Extraction of individual genome DNA of Chinese prawns
[0022] Take about 0.1g of Chinese prawn muscle tissue in a 2.0ml centrifuge tube, add 700μL of tissue lysate, add grinding beads, and homogenize the sample with a tissue homogenizer; slowly add 30μL of proteinase K with a concentration of 20mg / mL, and place in a 60°C water bath 3h, shake once every 30min, cool to room temperature after clarification; add 700μL of phenol-chloroform, shake well, centrifuge at 12000rpm for 15min; take supernatant, add 600μL of phenol-chloroform isoamyl alcohol (25:24:1), shake well , centrifuge at 12000rpm for 15min; take the supernatant and add 500μL of isopropanol, centrifuge at 12000rpm for 10min; pick out the DNA and wash it with 70% ethanol, dry it at room temperature, dilute it with sterilized water to 50ng / μL, and store it at -20℃ for later use.
Embodiment 2
[0023] Embodiment 2: Design of SNP marker amplification primers
[0024] Based on the FC1516SNP core sequence contained in the Chinese prawn transcriptome library, specific primers were designed at both ends of it to amplify the DNA fragment at this site. The specific primer sequences at both ends of the SNP core sequence of the present invention are: forward primer: 5'-ACCAATGGTCTCCCCCTTCC-3'; reverse primer sequence: 5'-AATCATCTCATCAGGACTGCTTG-3'.
[0025] The PCR amplification system is: Chinese shrimp genomic DNA 100ng, 10×PCR Buffer 2.5μL, 25mM MgCl 21.5μL, 2.5mM dNTP 2μL, 10μM positive and negative primers 2μL each; add sterilized water to 25μL; PCR reaction program: 94°C denaturation 5min, 95°C for 30sec, 60°C for 30sec, 72°C for 40sec, 30 cycles; 72°C for 10min. The PCR reaction program was: denaturation at 94°C for 5 min, 30 cycles at 95°C for 30 sec, 60°C for 30 sec, 72°C for 40 sec, and 72°C for 10 min.
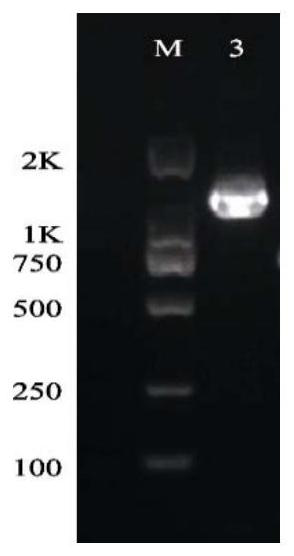
[0026] After the specificity of the PCR amplification produ...
Embodiment 3
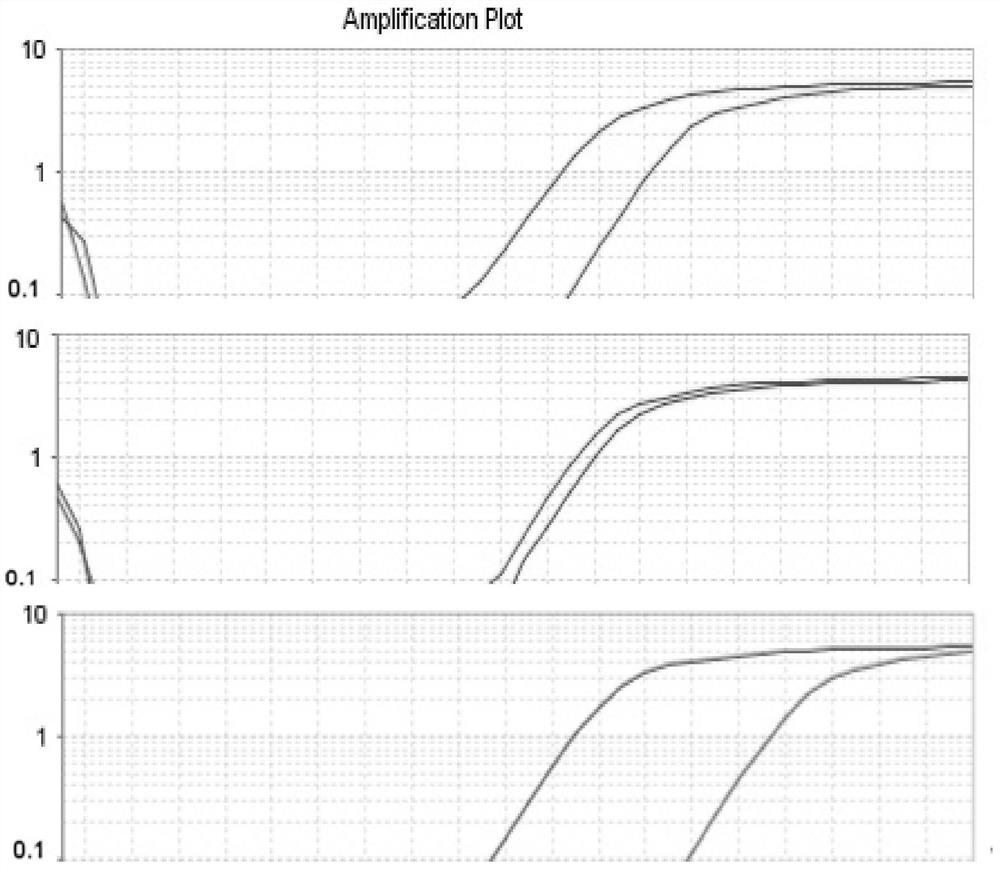
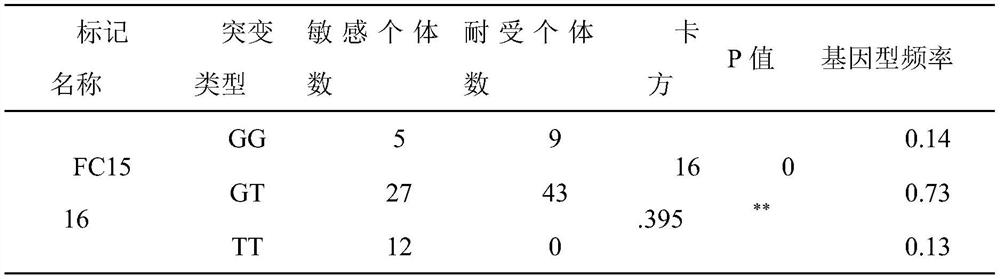
[0028] Example 3: Association analysis of Chinese prawn SNP markers and high pH tolerance traits
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com