CRISPR/Cas9 system for constructing Leber congenital amaurosis cloned porcine nuclear donor cells and application of CRISPR/Cas9 system
A technology for recombinant cells and expression vectors, applied in applications, nucleic acid vectors, animal cells, etc., can solve the problems of low probability of homozygous mutant offspring and inapplicability, and achieve improved nuclear localization capabilities, reduced workload, and multiplication speed slow effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 Construction of Cas9 high-efficiency expression vector and construction of pKG-U6gRNA vector
[0077] 1.1 Construction of pU6gRNA-eEF1a-mNLS-hSpCas9-EGFP-PURO (Cas9 high-efficiency expression vector)
[0078] (1) Remove redundant and invalid sequences in the gRNA backbone
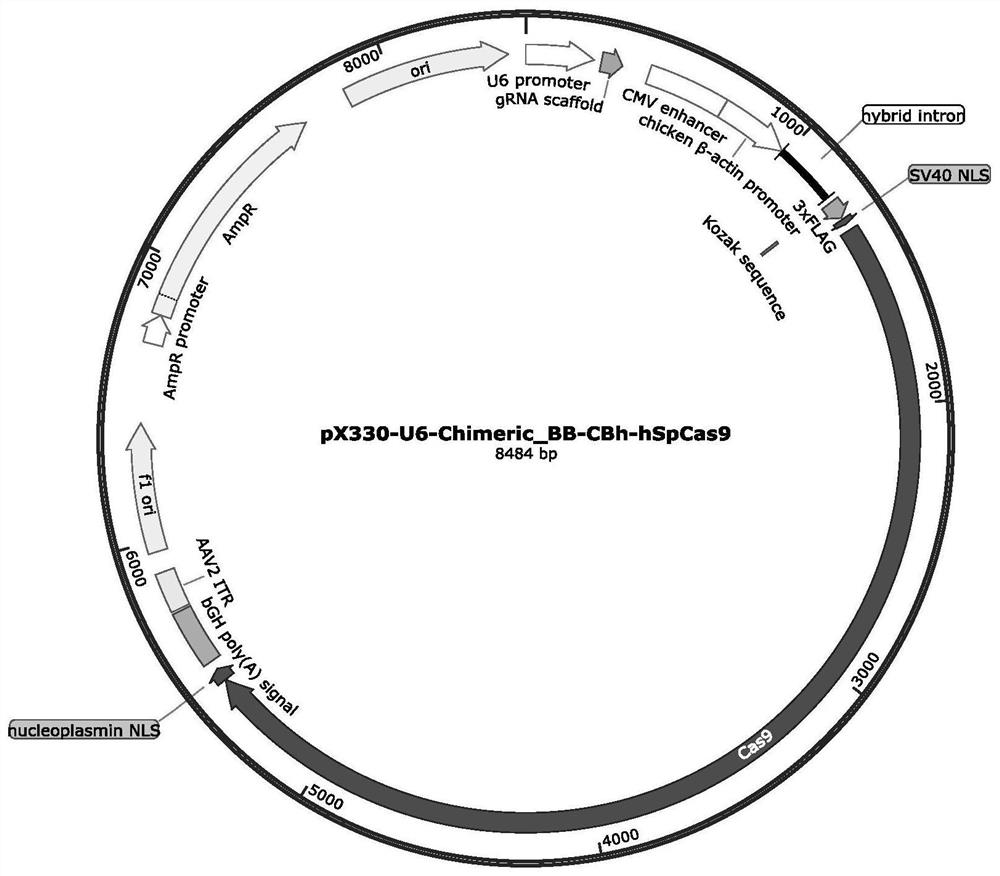
[0079] pX330-U6-Chimeric_BB-CBh-hSpCas9 (referred to as pX330, figure 1 ) was digested with BbsI and XbaI, and the vector fragment (about 8313bp) was recovered, and the insert fragment 175bp (SEQ ID NO: 31) was synthesized by the multi-fragment recombination method, and recombined with the recovered vector fragment to obtain the pU6gRNACas9 vector ( figure 2 ).
[0080] (2) Transformation of promoters and enhancers
[0081] For the constructed pU6gRNACas9 vector, use XbaI and AgeI endonucleases to remove the promoter (chickenβ-actin promoter) and enhancer sequence (CMV enhancer), recover the linear vector sequence of about 7650bp, and use the multi-fragment recombination method to synth...
Embodiment 2
[0096] Example 2 Plasmid Proportion Optimization and Effect Comparison of Plasmid pX330 and Plasmid pKG-GE3
[0097] 2.1 gRNA target design and construction
[0098] 2.1.1 Using Benchling to design gRNA targets for the RAG1 gene
[0099] RAG1-gRNA4: AGTTATGGCAGAACTCAGTG (SEQ ID NO.4)
[0100] The complementary DNA oligo of the inserted sequence of the synthetic RAG1 gene with a total of 2 targets is as follows:
[0101] RAG1-gRNA4S: caccgAGTTATGGCAGAACTCAGTG (SEQ ID NO.5)
[0102] RAG1-gRNA4A: aaacCACTGAGTTCTGCCATAACTc (SEQ ID NO. 6)
[0103] Both RAG1-gRNA4S and RAG1-gRNA4A are single-stranded DNA molecules.
[0104] 2.1.2 Primers designed to amplify the fragment containing the RAG1 gRNA target
[0105] RAG1-nF126: CCCCATCCAAAGTTTTTAAAGGA (SEQ ID NO. 7)
[0106] RAG1-nR525: TGTGGCAGATGTCACAGTTTAGG (SEQ ID NO. 8)
[0107] 2.1.3 The method of cloning the gRNA sequence into the pKG-U6gRNA backbone vector
[0108] 1) Digest 1ug pKG-U6gRNA plasmid with restriction endonucl...
Embodiment 3
[0158] Example 3 Design and construction of RPE65 gene target
[0159] 3.1 Genomic DNA extraction
[0160] 18 pigs (male A, B, C, D, E, F, G, H female 1, 2, 3, 4, 5) were performed using Vazyme's FastPure Cell / Tissue DNA Isolation Mini Kit (VazymeCat.DC102-01). , 6, 7, 8, 9, 10) Genomic DNA from ear tissue was extracted by column, quantified using NanoDrop, and stored at -20°C for future use.
[0161] 3.2 RPE65 gene knockout predetermined target and conservation analysis of adjacent genome sequences
[0162] 3.2.1 Pig RPE65 gene information
[0163] Encodes retinoid isomerohydrolase (Retinoid isomerohydrolase); located on chromosome 6; GeneID is 100516743, Sus scrofa. Existing research results have shown that RPE65 plays a central role in bone mass regulation. In pig genomic DNA, the RPE65 gene has 14 exons, of which the fifth exon occupies an important position in all transcripts (porcine RPE65 gene The 5th exon sequence, including the 4th exon, the 4th intron and part of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com