SNP sites remarkably associated with wheat powdery mildew resistance and application thereof in genetic breeding
A technology of wheat powdery mildew and loci, which is applied in the application field of genetics and molecular breeding, can solve the problems of high cost of deep sequencing, difficulty for breeders to master and use, and achieve the effect of low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Identification of SNPs by Simplified Genome Sequencing
[0036] 1.1 Sequencing materials
[0037] 768 wheat cultivars (lines) were selected for simplified genome sequencing to identify SNPs. These wheat varieties (lines) are mainly from the main wheat producing areas in my country, including the Huanghuai wheat area, the northern winter wheat area, the middle and lower reaches of the Yangtze River wheat area and the southwest wheat area.
[0038] Research methods
[0039] 1.2.1 Extraction of wheat genomic DNA
[0040]Seedling leaves were used to provide genomic DNA. DNA was extracted using the improved CTAB method. Specific operation: Take young wheat leaves in a 2mL centrifuge tube, freeze them with liquid nitrogen and grind them into powder on a tissue grinder; (b) add 800 μL CTAB to the 2mL tube, place in a 65°C water bath for 90 minutes, and gently Shake 5-8 times to fully lyse the DNA; (c) add 800 μL chloroform isoamyl alcohol (volume ratio 24:1) and...
Embodiment 2
[0049] Example 2 Genome-wide Identification of Powdery Mildew Resistance Gene Loci
[0050] 2.1 Materials
[0051] 768 accessions of wheat varieties (lines) from the main wheat producing areas in my country, including the Huanghuai wheat area, the northern winter wheat area, the middle and lower reaches of the Yangtze River wheat area, and the southwest wheat area, are the same as in Example 1.
[0052] 2.2 Method
[0053] 2.2.1 Identification of powdery mildew resistance
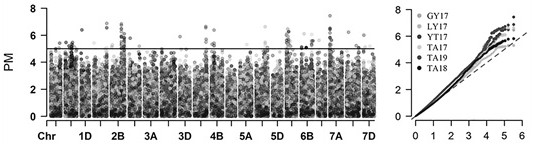
[0054] In order to identify the resistance of materials to powdery mildew, these 768 wheat varieties (lines) were planted in the experimental station of Shandong Agricultural University in Tai'an in 2017-2018 (TA17), 2018-2019 (TA18), and 2019-2020 (TA19) The disease identification nursery is used for identification and identification of powdery mildew resistance by artificial inoculation. At the same time, these materials were also planted in Yantai (YT), Luoyang (LY) and Guiyang (GY) in 2017-2018, and ...
Embodiment 3
[0068] Example 3 Wheat Powdery Mildew Resistance Gene qPm6A.3 verification and mapping
[0069] 3.1 Materials
[0070] The research materials include the powdery mildew-resistant parent 2013BP24, the susceptible parent Xumai 32 and the F 6 There are 126 recombinant inbred lines in total in the group of recombinant inbred lines. Huixianhong and Mingxian 169 were used as susceptible controls.
[0071] 3.2 Method
[0072] 3.2.1 Phenotype identification of disease resistance
[0073] With embodiment 1, the identification method of powdery mildew resistance
[0074] 3.2.2 DNA extraction
[0075] With embodiment 1.
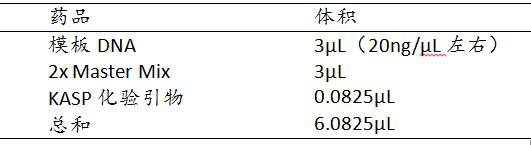
[0076] 3.2.3 KASP primer design and dilution:
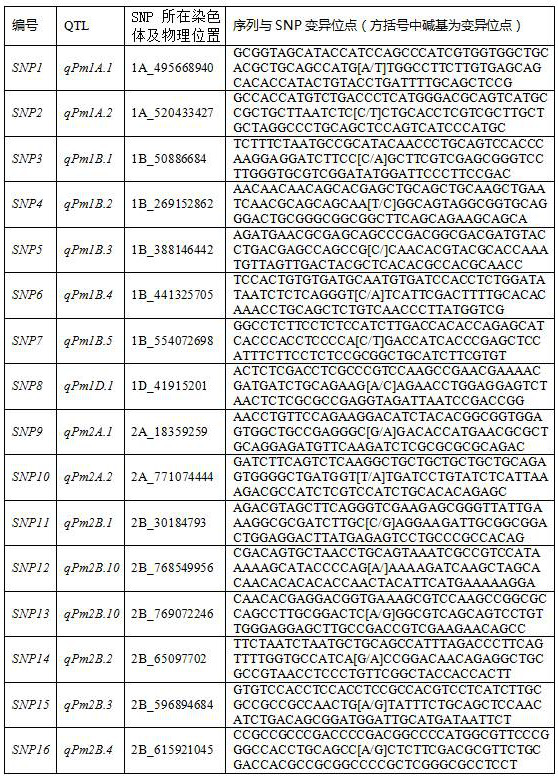
[0077]According to the results of GWAS analysis, the powdery mildew resistance QTL loci qPm6A.3 According to the significantly associated SNP (6A_86486561), a set (a total of three) of KASP primers k6A86486 was designed according to the SNP site information. Dilute the three primers of this set of primers to 10...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com